Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia
- PMID: 33831349
- PMCID: PMC8141024
- DOI: 10.1016/j.neuron.2021.03.024
Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia
Abstract
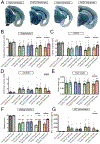
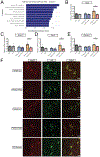
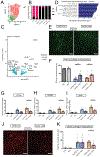
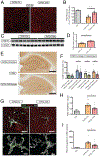
The apolipoprotein E (APOE) gene is the strongest genetic risk factor for Alzheimer's disease and directly influences tauopathy and tau-mediated neurodegeneration. ApoE4 has strong deleterious effects on both parameters. In the brain, apoE is produced and secreted primarily by astrocytes and by activated microglia. The cell-specific role of each form of apoE in the setting of neurodegeneration has not been determined. We generated P301S Tau/Aldh1l1-CreERT2/apoE3flox/flox or Tau/Aldh1l1-CreERT2/apoE4flox/flox mice. At 5.5 months of age, after the onset of tau pathology, we administered tamoxifen or vehicle and compared mice at 9.5 months of age. Removing astrocytic APOE4 markedly reduced tau-mediated neurodegeneration and decreased phosphorylated tau (pTau) pathology. Single-nucleus RNA sequencing analysis revealed striking gene expression changes in all cell types, with astrocytic APOE4 removal decreasing disease-associated gene signatures in neurons, oligodendrocytes, astrocytes, and microglia. Removal of astrocytic APOE4 decreased tau-induced synaptic loss and microglial phagocytosis of synaptic elements, suggesting a key role for astrocytic apoE in synaptic degeneration.
Keywords: APOE; astrocyte; microglia; neurodegeneration; tau.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.M.H. is as an inventor on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. C2N Diagnostics has licensed certain anti-tau antibodies to AbbVie for therapeutic development. D.M.H. is on the scientific advisory board of Denali and consults for Genentech, Merck, and Cajal Neuroscience. All other authors declare no competing interests.
Figures




Comment in
-
Astrocytic apoE4 and tau: Deadly combination for neurons.Cell Rep Med. 2021 Jun 15;2(6):100316. doi: 10.1016/j.xcrm.2021.100316. eCollection 2021 Jun 15. Cell Rep Med. 2021. PMID: 34195681 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

