Identification of SARS-CoV-2 viral entry inhibitors using machine learning and cell-based pseudotyped particle assay
- PMID: 33831697
- PMCID: PMC7997310
- DOI: 10.1016/j.bmc.2021.116119
Identification of SARS-CoV-2 viral entry inhibitors using machine learning and cell-based pseudotyped particle assay
Abstract
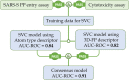
In response to the pandemic caused by SARS-CoV-2, we constructed a hybrid support vector machine (SVM) classification model using a set of publicly posted SARS-CoV-2 pseudotyped particle (PP) entry assay repurposing screen data to identify novel potent compounds as a starting point for drug development to treat COVID-19 patients. Two different molecular descriptor systems, atom typing descriptors and 3D fingerprints (FPs), were employed to construct the SVM classification models. Both models achieved reasonable performance, with the area under the curve of receiver operating characteristic (AUC-ROC) of 0.84 and 0.82, respectively. The consensus prediction outperformed the two individual models with significantly improved AUC-ROC of 0.91, where the compounds with inconsistent classifications were excluded. The consensus model was then used to screen the 173,898 compounds in the NCATS annotated and diverse chemical libraries. Of the 255 compounds selected for experimental confirmation, 116 compounds exhibited inhibitory activities in the SARS-CoV-2 PP entry assay with IC50 values ranged between 0.17 µM and 62.2 µM, representing an enrichment factor of 3.2. These 116 active compounds with diverse and novel structures could potentially serve as starting points for chemistry optimization for COVID-19 drug discovery.
Keywords: COVID-19; Consensus prediction; Pseudotyped particles assay; SARS-CoV-2; Support vector machine (SVM).
Published by Elsevier Ltd.
Conflict of interest statement
The authors declared that there is no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

