Whole-exome sequencing reveals the etiology of the rare primary hepatic mucoepidermoid carcinoma
- PMID: 33832503
- PMCID: PMC8034126
- DOI: 10.1186/s13000-021-01086-3
Whole-exome sequencing reveals the etiology of the rare primary hepatic mucoepidermoid carcinoma
Abstract
Background: Primary hepatic mucoepidermoid carcinoma (HMEC) is extremely rare and the molecular etiology is still unknown. The CRTC1-MAML2 fusion gene was previously detected in a primary HMEC, which is often associated with MEC of salivary gland in the literature.
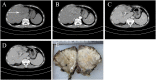
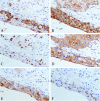
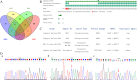
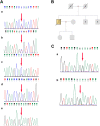
Methods: A 64-year-old male was diagnosed with HMEC based on malignant squamous cells and mucus-secreting cells in immunohistochemical examination. Fluorescence in situ hybridization (FISH) was used to detect the CRTC1-MAML2 fusion gene in HMEC. Whole-exome sequencing and Sanger sequencing were used to reveal the molecular characteristics of HMEC and analysis was performed with public data. Pedigree investigation was performed to identify susceptibility genes.
Results: Hematoxylin-eosin staining and immunohistochemistry revealed that the tumor cells were composed of malignant epidermoid malignant cells and mucous cells, indicating a diagnosis of HMEC. The CRTC1-MAML2 fusion gene was not detected in the primary HMEC, and somatic mutations in GNAS, KMT2C and ELF3 genes were identified by sequencing. Analyses of public data revealed somatic GNAS alterations in 2.1% hepatobiliary tumors and relation with parasite infection. Heterozygous germline mutations of FANCA, FANCI, FANCJ/BRIP1 and FAN1 genes were also identified. Pedigree investigation verified that mutation of Fanconi's anemia susceptibility genes were present in the pedigree.
Conclusions: Here we provide the first evidence of the molecular etiology of a rare HMEC associated with germline Fanconi's anemia gene mutations and somatic GNAS R201H mutation.
Keywords: Germline Fanconi’s anemia mutation; Hepatic mucoepidermoid carcinoma (HMEC); Somatic GNAS R201 mutation; Whole exome-sequencing (WES).
Conflict of interest statement
The authors declare no conflict of interests. The authors declare no competing financial interest.
Figures


References
-
- Tonon G, Modi S, Wu L, Kubo A, Coxon AB, Komiya T, O’Neil K, Stover K, el-Naggar A, Griffin JD, Kirsch IR, Kaye FJ. t(11;19)(q21;p13) translocation in mucoepidermoid carcinoma creates a novel fusion product that disrupts a Notch signaling pathway. Nat Genet. 2003;33(2):208–213. doi: 10.1038/ng1083. - DOI - PubMed
-
- Saeki K, Ohishi Y, Matsuda R, Mochidome N, Miyasaka Y, Yamamoto H, Koga Y, Maehara Y, Nakamura M, Oda Y. “Pancreatic mucoepidermoid carcinoma” is not a pancreatic counterpart of CRTC1/3-MAML2 fusion gene-related mucoepidermoid carcinoma of the salivary gland, and may more appropriately be termed pancreatic adenosquamous carcinoma with mucoepidermoid carcinoma-like features. Am J Surg Pathol. 2018;42(11):1419–1428. doi: 10.1097/PAS.0000000000001135. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

