Establishment of bovine expanded potential stem cells
- PMID: 33833056
- PMCID: PMC8053967
- DOI: 10.1073/pnas.2018505118
Establishment of bovine expanded potential stem cells
Abstract
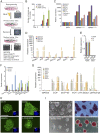
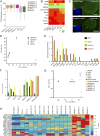
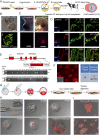
Embryonic stem cells (ESCs) and induced pluripotent stem cells have the potential to differentiate to all cell types of an adult individual and are useful for studying development and for translational research. However, extrapolation of mouse and human ESC knowledge to deriving stable ESC lines of domestic ungulates and large livestock species has been challenging. In contrast to ESCs that are usually established from the blastocyst, mouse expanded potential stem cells (EPSCs) are derived from four-cell and eight-cell embryos. We have recently used the EPSC approach and established stem cells from porcine and human preimplantation embryos. EPSCs are molecularly similar across species and have broader developmental potential to generate embryonic and extraembryonic cell lineages. We further explore the EPSC technology for mammalian species refractory to the standard ESC approaches and report here the successful establishment of bovine EPSCs (bEPSCs) from preimplantation embryos of both wild-type and somatic cell nuclear transfer. bEPSCs express high levels of pluripotency genes, propagate robustly in feeder-free culture, and are genetically stable in long-term culture. bEPSCs have enriched transcriptomic features of early preimplantation embryos and differentiate in vitro to cells of the three somatic germ layers and, in chimeras, contribute to both the embryonic (fetal) and extraembryonic cell lineages. Importantly, precise gene editing is efficiently achieved in bEPSCs, and genetically modified bEPSCs can be used as donors in somatic cell nuclear transfer. bEPSCs therefore hold the potential to substantially advance biotechnology and agriculture.
Keywords: bovine; expanded potential stem cell; nuclear transfer.
Copyright © 2021 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

References
-
- Evans M. J., Kaufman M. H., Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154–156 (1981). - PubMed
-
- Buehr M., et al. ., Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287–1298 (2008). - PubMed
-
- Thomson J. A., et al. ., Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

