Defective dystrophic thymus determines degenerative changes in skeletal muscle
- PMID: 33833239
- PMCID: PMC8032677
- DOI: 10.1038/s41467-021-22305-x
Defective dystrophic thymus determines degenerative changes in skeletal muscle
Abstract
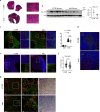
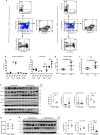
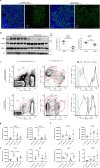
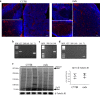
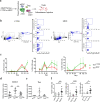
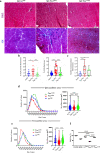
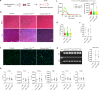
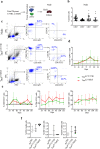
In Duchenne muscular dystrophy (DMD), sarcolemma fragility and myofiber necrosis produce cellular debris that attract inflammatory cells. Macrophages and T-lymphocytes infiltrate muscles in response to damage-associated molecular pattern signalling and the release of TNF-α, TGF-β and interleukins prevent skeletal muscle improvement from the inflammation. This immunological scenario was extended by the discovery of a specific response to muscle antigens and a role for regulatory T cells (Tregs) in muscle regeneration. Normally, autoimmunity is avoided by autoreactive T-lymphocyte deletion within thymus, while in the periphery Tregs monitor effector T-cells escaping from central regulatory control. Here, we report impairment of thymus architecture of mdx mice together with decreased expression of ghrelin, autophagy dysfunction and AIRE down-regulation. Transplantation of dystrophic thymus in recipient nude mice determine the up-regulation of inflammatory/fibrotic markers, marked metabolic breakdown that leads to muscle atrophy and loss of force. These results indicate that involution of dystrophic thymus exacerbates muscular dystrophy by altering central immune tolerance.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

