Mitochondrial NADP+ is essential for proline biosynthesis during cell growth
- PMID: 33833463
- PMCID: PMC9210447
- DOI: 10.1038/s42255-021-00374-y
Mitochondrial NADP+ is essential for proline biosynthesis during cell growth
Abstract
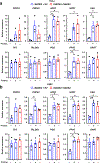
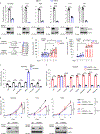
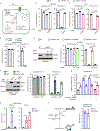
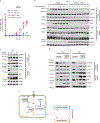
Nicotinamide adenine dinucleotide phosphate (NADP+) is vital to produce NADPH, a principal supplier of reducing power for biosynthesis of macromolecules and protection against oxidative stress. NADPH exists in separate pools, in both the cytosol and mitochondria; however, the cellular functions of mitochondrial NADPH are incompletely described. Here, we find that decreasing mitochondrial NADP(H) levels through depletion of NAD kinase 2 (NADK2), an enzyme responsible for production of mitochondrial NADP+, renders cells uniquely proline auxotrophic. Cells with NADK2 deletion fail to synthesize proline, due to mitochondrial NADPH deficiency. We uncover the requirement of mitochondrial NADPH and NADK2 activity for the generation of the pyrroline-5-carboxylate metabolite intermediate as the bottleneck step in the proline biosynthesis pathway. Notably, after NADK2 deletion, proline is required to support nucleotide and protein synthesis, making proline essential for the growth and proliferation of NADK2-deficient cells. Thus, we highlight proline auxotrophy in mammalian cells and discover that mitochondrial NADPH is essential to enable proline biosynthesis.
Conflict of interest statement
Competing interests
R.J.D. is an advisor for Agios Pharmaceuticals and Vida Ventures. All other authors declare no competing interests.
Figures
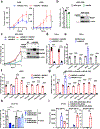

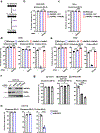


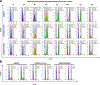
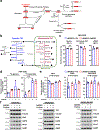





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

