Amino Acid Conjugates of Aminothiazole and Aminopyridine as Potential Anticancer Agents: Synthesis, Molecular Docking and in vitro Evaluation
- PMID: 33833504
- PMCID: PMC8021256
- DOI: 10.2147/DDDT.S297013
Amino Acid Conjugates of Aminothiazole and Aminopyridine as Potential Anticancer Agents: Synthesis, Molecular Docking and in vitro Evaluation
Abstract
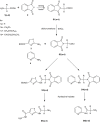
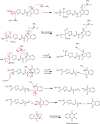
Purpose: The development of resistance to available anticancer drugs is increasingly becoming a major challenge and new chemical entities could be unveiled to compensate this therapeutic failure. The current study demonstrated the synthesis of 2-aminothiazole [S3(a-d) and S5(a-d)] and 2-aminopyridine [S4(a-d) and S6(a-d)] derivatives that can target multiple cellular networks implicated in cancer development.
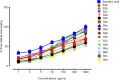
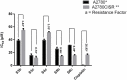
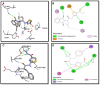
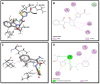
Methods: Biological assays were performed to investigate the antioxidant and anticancer potential of synthesized compounds. Redox imbalance and oxidative stress are hallmarks of cancer, therefore, synthesized compounds were preliminarily screened for their antioxidant activity using DPPH assay, and further five derivatives S3b, S3c, S4c, S5b, and S6c, with significant antioxidant potential, were selected for investigation of in vitro anticancer potential. The cytotoxic activities were evaluated against the parent (A2780) and cisplatin-resistant (A2780CISR) ovarian cancer cell lines. Further, Molecular docking studies of active compounds were performed to determine binding affinities.
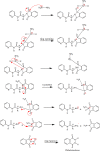
Results: Results revealed that S3c, S5b, and S6c displayed promising inhibition in cisplatin-resistant cell lines in comparison to parent cells in terms of both resistance factor (RF) and IC50 values. Moreover, S3c proved to be most active compound in both parent and resistant cell lines with IC50 values 15.57 µM and 11.52 µM respectively. Our docking studies demonstrated that compounds S3c, S5b, and S6c exhibited significant binding affinity with multiple protein targets of the signaling cascade.
Conclusion: Anticancer activities of compounds S3c, S5b, and S6c in cisplatin-resistant cell lines suggested that these ligands may contribute as lead compounds for the development of new anticancer drugs.
Keywords: anticancer activity; antioxidant activity; molecular docking; pyridine; thiazole.
© 2021 Naz et al.
Conflict of interest statement
The authors report no conflicts of interest for this work.
Figures


Similar articles
-
Synthesis and biological evaluation of purine-pyrazole hybrids incorporating thiazole, thiazolidinone or rhodanine moiety as 15-LOX inhibitors endowed with anticancer and antioxidant potential.Bioorg Chem. 2019 Jun;87:821-837. doi: 10.1016/j.bioorg.2019.03.076. Epub 2019 Apr 1. Bioorg Chem. 2019. PMID: 30999135
-
Molecular Dynamics and Biological Evaluation of 2-chloro-7-cyclopentyl- 7H-pyrrolo[2,3-d]pyrimidine Derivatives Against Breast Cancer.Comb Chem High Throughput Screen. 2017;20(8):703-712. doi: 10.2174/1386207320666170724110015. Comb Chem High Throughput Screen. 2017. PMID: 28738766
-
Molecular Docking, Antioxidant, Anticancer and Antileishmanial Effects of Newly Synthesized Quinoline Derivatives.Anticancer Agents Med Chem. 2020;20(13):1516-1529. doi: 10.2174/1871520620666200516145117. Anticancer Agents Med Chem. 2020. PMID: 32416701
-
Novel benzothiazole-based dual VEGFR-2/EGFR inhibitors targeting breast and liver cancers: Synthesis, cytotoxic activity, QSAR and molecular docking studies.Bioorg Med Chem Lett. 2022 Feb 15;58:128529. doi: 10.1016/j.bmcl.2022.128529. Epub 2022 Jan 7. Bioorg Med Chem Lett. 2022. PMID: 35007724 Review.
-
An Overview on Synthetic 2-Aminothiazole-Based Compounds Associated with Four Biological Activities.Molecules. 2021 Mar 7;26(5):1449. doi: 10.3390/molecules26051449. Molecules. 2021. PMID: 33800023 Free PMC article. Review.
Cited by
-
Cracking the code: the clinical and molecular impact of aminopyridines; a review (2019-2024).RSC Adv. 2025 Jan 8;15(1):688-711. doi: 10.1039/d4ra07438f. eCollection 2025 Jan 2. RSC Adv. 2025. PMID: 39781020 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

