Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients
- PMID: 33833618
- PMCID: PMC8018221
- DOI: 10.1155/2021/6635925
Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients
Abstract
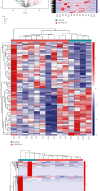
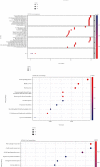
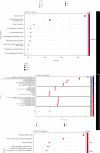
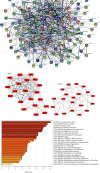
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was initially identified in China and currently worldwide dispersed, resulting in the coronavirus disease 2019 (COVID-19) pandemic. Notably, COVID-19 is characterized by systemic inflammation. However, the potential mechanisms of the "cytokine storm" of COVID-19 are still limited. In this study, fourteen peripheral blood samples from COVID-19 patients (n = 10) and healthy donors (n = 4) were collected to perform the whole-transcriptome sequencing. Lung tissues of COVID-19 patients (70%) presenting with ground-glass opacity. Also, the leukocytes and lymphocytes were significantly decreased in COVID-19 compared with the control group (p < 0.05). In total, 25,482 differentially expressed messenger RNAs (DE mRNA), 23 differentially expressed microRNAs (DE miRNA), and 410 differentially expressed long noncoding RNAs (DE lncRNAs) were identified in the COVID-19 samples compared to the healthy controls. Gene Ontology (GO) analysis showed that the upregulated DE mRNAs were mainly involved in antigen processing and presentation of endogenous antigen, positive regulation of T cell mediated cytotoxicity, and positive regulation of gamma-delta T cell activation. The downregulated DE mRNAs were mainly concentrated in the glycogen biosynthetic process. We also established the protein-protein interaction (PPI) networks of up/downregulated DE mRNAs and identified 4 modules. Functional enrichment analyses indicated that these module targets were associated with positive regulation of cytokine production, cytokine-mediated signaling pathway, leukocyte differentiation, and migration. A total of 6 hub genes were selected in the PPI module networks including AKT1, TNFRSF1B, FCGR2A, CXCL8, STAT3, and TLR2. Moreover, a competing endogenous RNA network showed the interactions between lncRNAs, mRNAs, and miRNAs. Our results highlight the potential pathogenesis of excessive cytokine production such as MSTRG.119845.30/hsa-miR-20a-5p/TNFRSF1B, MSTRG.119845.30/hsa-miR-29b-2-5p/FCGR2A, and MSTRG.106112.2/hsa-miR-6501-5p/STAT3 axis, which may also play an important role in the development of ground-glass opacity in COVID-19 patients. This study gives new insights into inflammation regulatory mechanisms of coding and noncoding RNAs in COVID-19, which may provide novel diagnostic biomarkers and therapeutic avenues for COVID-19 patients.
Copyright © 2021 Cai-xia Li et al.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Worldometer COVID-19 Coronavirus Pandemic. 2021. https://www.worldometers.info/coronavirus/
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

