Metagenomic Data Assembly - The Way of Decoding Unknown Microorganisms
- PMID: 33833738
- PMCID: PMC8021871
- DOI: 10.3389/fmicb.2021.613791
Metagenomic Data Assembly - The Way of Decoding Unknown Microorganisms
Abstract
Metagenomics is a segment of conventional microbial genomics dedicated to the sequencing and analysis of combined genomic DNA of entire environmental samples. The most critical step of the metagenomic data analysis is the reconstruction of individual genes and genomes of the microorganisms in the communities using metagenomic assemblers - computational programs that put together small fragments of sequenced DNA generated by sequencing instruments. Here, we describe the challenges of metagenomic assembly, a wide spectrum of applications in which metagenomic assemblies were used to better understand the ecology and evolution of microbial ecosystems, and present one of the most efficient microbial assemblers, SPAdes that was upgraded to become applicable for metagenomics.
Keywords: SPAdes; algorithms; metagenomic assembly; metagenomics; microbiota.
Copyright © 2021 Lapidus and Korobeynikov.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
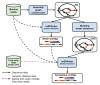
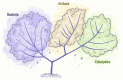

References
-
- Andrews S., Krueger F., Segonds-Pichon A., Biggins L., Krueger C., Wingett S., et al. (2010). FastQC: a quality control tool for high throughput sequence data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (Accessed March 10, 2021).
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

