Immune-Inhibitory Gene Expression is Positively Correlated with Overall Immune Activity and Predicts Increased Survival Probability of Cervical and Head and Neck Cancer Patients
- PMID: 33834038
- PMCID: PMC8021786
- DOI: 10.3389/fmolb.2021.622643
Immune-Inhibitory Gene Expression is Positively Correlated with Overall Immune Activity and Predicts Increased Survival Probability of Cervical and Head and Neck Cancer Patients
Abstract
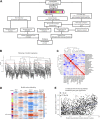
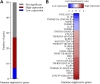
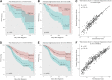
Background: Limited immunotherapy options are approved for the treatment of cervical cancer and only 10-25% of patients respond effectively to checkpoint inhibition monotherapy. To aid the development of novel therapeutic immune targets, we aimed to explore survival-associated immune biomarkers and co-expressed immune networks in cervical cancer. Methods: Using The Cancer Genome Atlas (TCGA) Cervical Squamous Cell Carcinoma (CESC) data (n = 304), we performed weighted gene co-expression network analysis (WGCNA), and determined which co-expressed immune-related genes and networks are associated with survival probability in CESC patients under conventional therapy. A "Pan-Immune Score" and "Immune Suppression Score" was generated based on expression of survival-associated co-expressed immune networks and immune suppressive genes, which were subsequently tested for association with survival probablity using the TCGA Head Neck Squamous Cell Carcinoma (HNSCC) data (n = 528), representing a second SCC cancer type. Results: In CESC, WGCNA identified a co-expression module enriched in immune response related genes, including 462 genes where high expression was associated with increased survival probability, and enriched for genes associated with T cell receptor, cytokine and chemokine signaling. However, a high level of expression of 43 of the genes in this module was associated with decreased survival probability but were not enriched in particular pathways. Separately, we identified 20 genes associated with immune suppression including inhibitory immune checkpoint and regulatory T cell-related genes, where high expression was associated with increased survival probability. Expression of these 20 immune suppressive genes (represented as "Immune Suppression Score") was highly correlated with expression of overall survival-associated immune genes (represented as "Pan-Immune Score"). However, high expression of seven immune suppression genes, including TWEAK-R, CD73, IL1 family and TGFb family genes, was significantly associated with decreased survival probability. Both scores also significantly associated with survival probability in HNSCC, and correlated with the previously established "Immunophenoscore." Conclusion: CESC and HNSCC tumors expressing genes predictive of T cell infiltrates (hot tumors) have a better prognosis, despite simultaneous expression of many immune inhibitory genes, than tumors lacking expression of genes associated with T cell infiltrates (cold tumors) whether or not these tumor express immune inhibitory genes.
Keywords: cervical cancer; head and neck cancer; immune checkpoints; immune inhibition; inflamed tumours; prognosis.
Copyright © 2021 Budhwani, Turrell, Yu, Frazer, Mehdi and Chandra.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

