Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs
- PMID: 33834049
- PMCID: PMC8021921
- DOI: 10.3389/fvets.2021.664718
Horizons in Veterinary Precision Oncology: Fundamentals of Cancer Genomics and Applications of Liquid Biopsy for the Detection, Characterization, and Management of Cancer in Dogs
Abstract
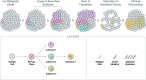
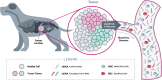
Cancer is the leading cause of death in dogs, in part because many cases are identified at an advanced stage when clinical signs have developed, and prognosis is poor. Increased understanding of cancer as a disease of the genome has led to the introduction of liquid biopsy testing, allowing for detection of genomic alterations in cell-free DNA fragments in blood to facilitate earlier detection, characterization, and management of cancer through non-invasive means. Recent discoveries in the areas of genomics and oncology have provided a deeper understanding of the molecular origins and evolution of cancer, and of the "one health" similarities between humans and dogs that underlie the field of comparative oncology. These discoveries, combined with technological advances in DNA profiling, are shifting the paradigm for cancer diagnosis toward earlier detection with the goal of improving outcomes. Liquid biopsy testing has already revolutionized the way cancer is managed in human medicine - and it is poised to make a similar impact in veterinary medicine. Multiple clinical use cases for liquid biopsy are emerging, including screening, aid in diagnosis, targeted treatment selection, treatment response monitoring, minimal residual disease detection, and recurrence monitoring. This review article highlights key scientific advances in genomics and their relevance for veterinary oncology, with the goal of providing a foundational introduction to this important topic for veterinarians. As these technologies migrate from human medicine into veterinary medicine, improved awareness and understanding will facilitate their rapid adoption, for the benefit of veterinary patients.
Keywords: cancer; cell-free DNA; cfDNA; circulating tumor DNA; dog; genomic; liquid biopsy; one health.
Copyright © 2021 Chibuk, Flory, Kruglyak, Leibman, Nahama, Dharajiya, van den Boom, Jensen, Friedman, Shen, Clemente-Vicario, Chorny, Tynan, Lytle, Holtvoigt, Murtaza, Diaz Jr., Tsui and Grosu.
Conflict of interest statement
JC, AF, KK, IC, JT, KL, LH, DT, and DG are employed by or affiliated with PetDx. JC, AF, KK, NL, AN, ND, DB, TJ, JF, MS, IC, JT, KL, LH, MM, LD, DT, and DG hold vested or unvested equity in PetDx. TJ is employed by Laboratory Corporation of America. JF is Managing Partner at Friedman Bioventure, Inc. MS is Managing Director at RS Technology Ventures LLC. KK is an inventor on multiple patent applications related to bioinformatics methods for cancer diagnostics and holds equity in Illumina. MM is an inventor on multiple patent applications covering technologies for canine and human cancer diagnostics, and has licensing or consulting relationships with PetDx, Exact Sciences, AstraZeneca, Bristol Myers Squibb, and TGen. LD is a member of the board of directors of Personal Genome Diagnostics (PGDx) and Jounce Therapeutics. LD is a compensated consultant to PGDx, 4Paws (PetDx), Innovatus CP, Se'er, Kinnate and Neophore. LD is an uncompensated consultant for Merck but has received research support for clinical trials from Merck. LD is an inventor of multiple licensed patents related to technology for circulating tumor DNA analyses and mismatch repair deficiency for diagnosis and therapy from Johns Hopkins University. Some of these licenses and relationships are associated with equity or royalty payments directly to Johns Hopkins and LD. LD holds equity in PGDx, Jounce Therapeutics, Thrive Earlier Detection, Se'er, Kinnate and Neophore. LD's spouse holds equity in Amgen. The terms of all these arrangements for LD are being managed by Johns Hopkins and Memorial Sloan Kettering in accordance with their conflict of interest policies. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

