BMP9-ID1 signaling promotes EpCAM-positive cancer stem cell properties in hepatocellular carcinoma
- PMID: 33834612
- PMCID: PMC8333780
- DOI: 10.1002/1878-0261.12963
BMP9-ID1 signaling promotes EpCAM-positive cancer stem cell properties in hepatocellular carcinoma
Abstract
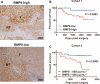
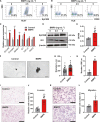
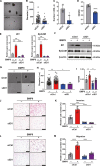
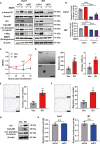
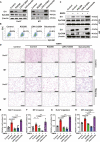
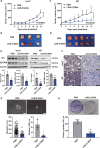
The malignant nature of hepatocellular carcinoma (HCC) is closely related to the presence of cancer stem cells (CSCs). Bone morphologic protein 9 (BMP9), a member of the transforming growth factor-beta (TGF-β) superfamily, was recently reported to be involved in liver diseases including cancer. We aimed to elucidate the role of BMP9 signaling in HCC-CSC properties and to assess the therapeutic effect of BMP receptor inhibitors in HCC. We have identified that high BMP9 expression in tumor tissues or serum from patients with HCC leads to poorer outcome. BMP9 promoted CSC properties in epithelial cell adhesion molecule (EpCAM)-positive HCC subtype via enhancing inhibitor of DNA-binding protein 1 (ID1) expression in vitro. Additionally, ID1 knockdown significantly repressed BMP9-promoted HCC-CSC properties by suppressing Wnt/β-catenin signaling. Interestingly, cells treated with BMP receptor inhibitors K02288 and LDN-212854 blocked HCC-CSC activation by inhibiting BMP9-ID1 signaling, in contrast to cells treated with the TGF-β receptor inhibitor galunisertib. Treatment with LDN-212854 suppressed HCC tumor growth by repressing ID1 and EpCAM in vivo. Our study demonstrates the pivotal role of BMP9-ID1 signaling in promoting HCC-CSC properties and the therapeutic potential of BMP receptor inhibitors in treating EpCAM-positive HCC. Therefore, targeting BMP9-ID1 signaling could offer novel therapeutic options for patients with malignant HCC.
Keywords: BMP receptor inhibitor; BMP9-ID1 signaling; EpCAM; cancer stem cells; hepatocellular carcinoma.
© 2021 The Authors. Molecular Oncology published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Elkhenany H, Shekshek A, Abdel‐Daim M & El‐Badri N (2020) Stem cell therapy for hepatocellular carcinoma: future perspectives. Adv Exp Med Biol 1237, 97–119. - PubMed
-
- Visvader JE & Lindeman GJ (2008) Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer 8, 755–768. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

