In-frame deletion in canine PITRM1 is associated with a severe early-onset epilepsy, mitochondrial dysfunction and neurodegeneration
- PMID: 33835239
- PMCID: PMC8519929
- DOI: 10.1007/s00439-021-02279-y
In-frame deletion in canine PITRM1 is associated with a severe early-onset epilepsy, mitochondrial dysfunction and neurodegeneration
Abstract
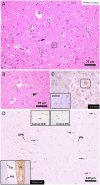
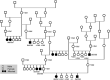
We investigated the clinical, genetic, and pathological characteristics of a previously unknown severe juvenile brain disorder in several litters of Parson Russel Terriers. The disease started with epileptic seizures at 6-12 weeks of age and progressed rapidly to status epilepticus and death or euthanasia. Histopathological changes at autopsy were restricted to the brain. There was severe acute neuronal degeneration and necrosis diffusely affecting the grey matter throughout the brain with extensive intraneuronal mitochondrial crowding and accumulation of amyloid-β (Aβ). Combined homozygosity mapping and genome sequencing revealed an in-frame 6-bp deletion in the nuclear-encoded pitrilysin metallopeptidase 1 (PITRM1) encoding for a mitochondrial protease involved in mitochondrial targeting sequence processing and degradation. The 6-bp deletion results in the loss of two amino acid residues in the N-terminal part of PITRM1, potentially affecting protein folding and function. Assessment of the mitochondrial function in the affected brain tissue showed a significant deficiency in respiratory chain function. The functional consequences of the mutation were modeled in yeast and showed impaired growth in permissive conditions and an impaired respiration capacity. Loss-of-function variants in human PITRM1 result in a childhood-onset progressive amyloidotic neurological syndrome characterized by spinocerebellar ataxia with behavioral, psychiatric and cognitive abnormalities. Homozygous Pitrm1-knockout mice are embryonic lethal, while heterozygotes show a progressive, neurodegenerative phenotype characterized by impairment in motor coordination and Aβ deposits. Our study describes a novel early-onset PITRM1-related neurodegenerative canine brain disorder with mitochondrial dysfunction, Aβ accumulation, and lethal epilepsy. The findings highlight the essential role of PITRM1 in neuronal survival and strengthen the connection between mitochondrial dysfunction and neurodegeneration.
© 2021. The Author(s).
Conflict of interest statement
HL is a paid consultant to Genoscoper Laboratories Ltd providing genetic tests for dogs. JD is an employee of Genoscoper Laboratories Ltd.
Figures







References
-
- Berendt M, Farquhar RG, Mandigers PJ, Pakozdy A, Bhatti SF, De Risio L, Fischer A, Long S, Matiasek K, Munana K, Patterson EE, Penderis J, Platt S, Podell M, Potschka H, Pumarola MB, Rusbridge C, Stein VM, Tipold A, Volk HA. International veterinary epilepsy task force consensus report on epilepsy definition, classification and terminology in companion animals. BMC Vet Res. 2015;11:182–192. doi: 10.1186/s12917-015-0461-2. - DOI - PMC - PubMed
-
- Broeckx BJ, Hitte C, Coopman F, Verhoeven GE, De Keulenaer S, De Meester E, Derrien T, Alfoldi J, Lindblad-Toh K, Bosmans T, Gielen I, Van Bree H, Van Ryssen B, Saunders JH, Van Nieuwerburgh F, Deforce D. Improved canine exome designs, featuring ncRNAs and increased coverage of protein coding genes. Sci Rep. 2015;5:12810. doi: 10.1038/srep12810. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

