Purification and characterization of thermoactive serratiopeptidase from Serratia marcescens AD-W2
- PMID: 33835269
- PMCID: PMC8035408
- DOI: 10.1186/s13568-021-01215-7
Purification and characterization of thermoactive serratiopeptidase from Serratia marcescens AD-W2
Erratum in
-
Correction to: Purification and characterization of thermoactive serratiopeptidase from Serratia marcescens AD-W2.AMB Express. 2021 May 4;11(1):64. doi: 10.1186/s13568-021-01223-7. AMB Express. 2021. PMID: 33948770 Free PMC article. No abstract available.
Abstract
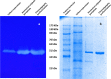
Serratiopeptidase is a proteolytic enzyme extensively used as an anti-inflammatory and analgesic drug. Present work reports a thermoactive serratiopeptidase from Serratia marcescens AD-W2, a soil isolate from the North-Western Himalayan region of India. The extracellular metalloprotease has been purified by a simple two-step procedure resulting in a specific activity of 20,492 Units/mg protein with 5.28-fold purification. The molecular mass of the metalloprotease, as determined by SDS-PAGE was ~ 51 kDa. The purified serratiopeptidase presented optimum activity at pH 9.0, temperature 50 °C and stability in wide pH and temperature range. Critical temperature of 50 °C confirmed the thermoactivity of the purified serratiopeptidase. The kinetic studies of the purified serratiopeptidase revealed Vmax and Km of 57,256 Units/mL and 1.57 mg/mL, respectively, for casein. The purified serratiopeptidase from S. marcescens AD-W2 was found to be 100% identical to serralysin from Serratia marcescens ATCC 21074/E-15. The catalytic domain comprising of Zn coordinated with three histidine residues (His192, His196, His202), along with glutamate (Glu193) and tyrosine (Tyr232) residues, further confirmed that the purified protein is identical to serralysin.
Keywords: Homology modelling; Metalloprotease; Serralysin; Serratia marcescens; Serratiopeptidase.
Conflict of interest statement
All the authors declare that they do not have any conflict of interest.
Figures








Similar articles
-
Serratiopeptidase: An integrated View of Multifaceted Therapeutic Enzyme.Biomolecules. 2022 Oct 13;12(10):1468. doi: 10.3390/biom12101468. Biomolecules. 2022. PMID: 36291677 Free PMC article. Review.
-
Purification and characterization of a metalloprotease produced by the C8 isolate of Serratia marcescens using silkworm pupae or casein as a protein source.Int J Biol Macromol. 2019 Aug 15;135:97-105. doi: 10.1016/j.ijbiomac.2019.05.122. Epub 2019 May 21. Int J Biol Macromol. 2019. PMID: 31125647
-
Purification, functional characterization and enhanced production of serratiopeptidase from Serratia marcescens MES-4: An endophyte isolated from Morus rubra.J Biotechnol. 2024 May 20;387:58-68. doi: 10.1016/j.jbiotec.2024.04.002. Epub 2024 Apr 4. J Biotechnol. 2024. PMID: 38582407
-
Production, purification and characterization of a 50-kDa extracellular metalloprotease from Serratia marcescens.Appl Microbiol Biotechnol. 1997 Sep;48(3):317-24. doi: 10.1007/s002530051056. Appl Microbiol Biotechnol. 1997. PMID: 9352674
-
Advances and challenges in serratiopeptidase topical formulation.Ann Pharm Fr. 2024 Nov;82(6):966-979. doi: 10.1016/j.pharma.2024.05.008. Epub 2024 May 29. Ann Pharm Fr. 2024. PMID: 38821483 Review.
Cited by
-
Scale-Up of the Fermentation Process for the Production and Purification of Serratiopeptidase Using Silkworm Pupae as a Substrate.Methods Protoc. 2024 Feb 25;7(2):19. doi: 10.3390/mps7020019. Methods Protoc. 2024. PMID: 38525777 Free PMC article.
-
Update of the list of qualified presumption of safety (QPS) recommended microbiological agents intentionally added to food or feed as notified to EFSA 19: Suitability of taxonomic units notified to EFSA until September 2023.EFSA J. 2024 Jan 11;22(1):e8517. doi: 10.2903/j.efsa.2024.8517. eCollection 2024 Jan. EFSA J. 2024. PMID: 38213415 Free PMC article.
-
Serratiopeptidase: An integrated View of Multifaceted Therapeutic Enzyme.Biomolecules. 2022 Oct 13;12(10):1468. doi: 10.3390/biom12101468. Biomolecules. 2022. PMID: 36291677 Free PMC article. Review.
-
Production of a Recombinant Fibrinolytic Protease from an Isolate of Serratia marcescens from the Amazon Basin.ACS Omega. 2025 May 14;10(20):20854-20865. doi: 10.1021/acsomega.5c02194. eCollection 2025 May 27. ACS Omega. 2025. PMID: 40454077 Free PMC article.
-
Increased Proteolytic Activity of Serratia marcescens Clinical Isolate HU1848 Is Associated with Higher eepR Expression.Pol J Microbiol. 2024 Mar 4;73(1):11-20. doi: 10.33073/pjm-2024-002. eCollection 2024 Mar 1. Pol J Microbiol. 2024. PMID: 38437469 Free PMC article.
References
-
- Devi CS, Joseph RE, Saravanan H, Naine SJ, Srinivansan VM. Screening and molecular characterization of Serratia marcescens VITSD2: a strain producing optimum serratiopeptidase. Front Biol. 2013;8:632–639. doi: 10.1007/s11515-013-1284-9. - DOI
-
- Ethiraj S, Gopinath S. Production, purification, characterization, immobilization, and application of serrapeptase: a review. Front Biol. 2017;12:333–348. doi: 10.1007/s11515-017-1461-3. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

