Using a Secure, Continually Updating, Web Source Processing Pipeline to Support the Real-Time Data Synthesis and Analysis of Scientific Literature: Development and Validation Study
- PMID: 33835932
- PMCID: PMC8104004
- DOI: 10.2196/25714
Using a Secure, Continually Updating, Web Source Processing Pipeline to Support the Real-Time Data Synthesis and Analysis of Scientific Literature: Development and Validation Study
Abstract
Background: The scale and quality of the global scientific response to the COVID-19 pandemic have unquestionably saved lives. However, the COVID-19 pandemic has also triggered an unprecedented "infodemic"; the velocity and volume of data production have overwhelmed many key stakeholders such as clinicians and policy makers, as they have been unable to process structured and unstructured data for evidence-based decision making. Solutions that aim to alleviate this data synthesis-related challenge are unable to capture heterogeneous web data in real time for the production of concomitant answers and are not based on the high-quality information in responses to a free-text query.
Objective: The main objective of this project is to build a generic, real-time, continuously updating curation platform that can support the data synthesis and analysis of a scientific literature framework. Our secondary objective is to validate this platform and the curation methodology for COVID-19-related medical literature by expanding the COVID-19 Open Research Dataset via the addition of new, unstructured data.
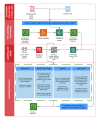
Methods: To create an infrastructure that addresses our objectives, the PanSurg Collaborative at Imperial College London has developed a unique data pipeline based on a web crawler extraction methodology. This data pipeline uses a novel curation methodology that adopts a human-in-the-loop approach for the characterization of quality, relevance, and key evidence across a range of scientific literature sources.
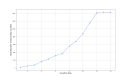
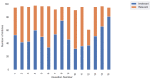
Results: REDASA (Realtime Data Synthesis and Analysis) is now one of the world's largest and most up-to-date sources of COVID-19-related evidence; it consists of 104,000 documents. By capturing curators' critical appraisal methodologies through the discrete labeling and rating of information, REDASA rapidly developed a foundational, pooled, data science data set of over 1400 articles in under 2 weeks. These articles provide COVID-19-related information and represent around 10% of all papers about COVID-19.
Conclusions: This data set can act as ground truth for the future implementation of a live, automated systematic review. The three benefits of REDASA's design are as follows: (1) it adopts a user-friendly, human-in-the-loop methodology by embedding an efficient, user-friendly curation platform into a natural language processing search engine; (2) it provides a curated data set in the JavaScript Object Notation format for experienced academic reviewers' critical appraisal choices and decision-making methodologies; and (3) due to the wide scope and depth of its web crawling method, REDASA has already captured one of the world's largest COVID-19-related data corpora for searches and curation.
Keywords: COVID-19; critical analysis; data; data science; data synthesis; database; decision making; infodemic; infrastructure; literature; methodology; misinformation; pipeline; research; structured data synthesis; web crawl data.
©Uddhav Vaghela, Simon Rabinowicz, Paris Bratsos, Guy Martin, Epameinondas Fritzilas, Sheraz Markar, Sanjay Purkayastha, Karl Stringer, Harshdeep Singh, Charlie Llewellyn, Debabrata Dutta, Jonathan M Clarke, Matthew Howard, PanSurg REDASA Curators, Ovidiu Serban, James Kinross. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 06.05.2021.
Conflict of interest statement
Conflicts of Interest: GM received equity from Medical iSight (Augmented Reality). SP provides consultations for Medtronic, T.M.L.E. Ltd., and Roche. SP is also the cofounder and director of Mangetoo, 1 World Medical, and the London General Surgery Clinic. SP is also a partner of One Welbeck Hospital. JK provides consultations for Verb robotics, Safeheal, YSOPIA bioscience, and Universal Diagnostics (UDX). JK also received equity from Mangetoo (teledietetics), 1 Welbeck Day Surgery (Hospital), 1 World medical (Personal Protective Equipment), and Medical iSight (Augmented Reality). The other authors have no conflicts of interest to declare for this paper.
Figures




References
-
- LitCovid. National Center for Biotechnology Information. [2021-04-12]. https://www.ncbi.nlm.nih.gov/research/coronavirus/
-
- COVID-19 research update: How many pandemic papers have been published? Nature Index. [2020-10-01]. https://www.natureindex.com/news-blog/how-coronavirus-is-changing-resear....
-
- COVID-19 rapid guideline: critical care in adults. National Institute for Health and Care Excellence. [2020-10-01]. https://www.nice.org.uk/guidance/ng159. - PubMed
-
- Interim process and methods for developing rapid guidelines on COVID-19. National Institute for Health and Care Excellence. [2020-10-01]. https://www.nice.org.uk/process/pmg35/chapter/scoping.
-
- Countering Missinformation about COVID-19. World Health Organization. [2020-10-01]. https://www.who.int/news-room/feature-stories/detail/countering-misinfor....
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

