Borealin directs recruitment of the CPC to oocyte chromosomes and movement to the microtubules
- PMID: 33836043
- PMCID: PMC8185691
- DOI: 10.1083/jcb.202006018
Borealin directs recruitment of the CPC to oocyte chromosomes and movement to the microtubules
Abstract
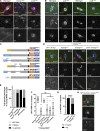
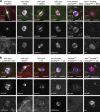
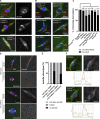
The chromosomes in the oocytes of many animals appear to promote bipolar spindle assembly. In Drosophila oocytes, spindle assembly requires the chromosome passenger complex (CPC), which consists of INCENP, Borealin, Survivin, and Aurora B. To determine what recruits the CPC to the chromosomes and its role in spindle assembly, we developed a strategy to manipulate the function and localization of INCENP, which is critical for recruiting the Aurora B kinase. We found that an interaction between Borealin and the chromatin is crucial for the recruitment of the CPC to the chromosomes and is sufficient to build kinetochores and recruit spindle microtubules. HP1 colocalizes with the CPC on the chromosomes and together they move to the spindle microtubules. We propose that the Borealin interaction with HP1 promotes the movement of the CPC from the chromosomes to the microtubules. In addition, within the central spindle, rather than at the centromeres, the CPC and HP1 are required for homologous chromosome bi-orientation.
© 2021 Wang et al.
Figures





Similar articles
-
The chromosomal passenger complex activates Polo kinase at centromeres.PLoS Biol. 2012 Jan;10(1):e1001250. doi: 10.1371/journal.pbio.1001250. Epub 2012 Jan 24. PLoS Biol. 2012. PMID: 22291575 Free PMC article.
-
Switching of INCENP paralogs controls transitions in mitotic chromosomal passenger complex functions.Cell Cycle. 2019 Sep;18(17):2006-2025. doi: 10.1080/15384101.2019.1634954. Epub 2019 Jul 15. Cell Cycle. 2019. PMID: 31306061 Free PMC article.
-
The Borealin dimerization domain interacts with Sgo1 to drive Aurora B-mediated spindle assembly.Mol Biol Cell. 2020 Sep 15;31(20):2207-2218. doi: 10.1091/mbc.E20-05-0341. Epub 2020 Jul 22. Mol Biol Cell. 2020. PMID: 32697622 Free PMC article.
-
The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis.Nat Rev Mol Cell Biol. 2012 Dec;13(12):789-803. doi: 10.1038/nrm3474. Nat Rev Mol Cell Biol. 2012. PMID: 23175282 Free PMC article. Review.
-
The Molecular Mechanism of Aurora-B Regulating Kinetochore-Microtubule Attachment in Mitosis and Oocyte Meiosis.Cytogenet Genome Res. 2024;164(2):69-77. doi: 10.1159/000540588. Epub 2024 Jul 27. Cytogenet Genome Res. 2024. PMID: 39068909 Review.
Cited by
-
Multiple pools of PP2A regulate spindle assembly, kinetochore attachments and cohesion in Drosophila oocytes.J Cell Sci. 2021 Jul 15;134(14):jcs254037. doi: 10.1242/jcs.254037. Epub 2021 Jul 23. J Cell Sci. 2021. PMID: 34297127 Free PMC article.
-
Highway to hell-thy meiotic divisions: Chromosome passenger complex functions driven by microtubules: CPC interactions with both the chromosomes and microtubules are important for spindle assembly and function: CPC interactions with both the chromosomes and microtubules are important for spindle assembly and function.Bioessays. 2022 Jan;44(1):e2100202. doi: 10.1002/bies.202100202. Epub 2021 Nov 25. Bioessays. 2022. PMID: 34821405 Free PMC article.
-
Genome-Wide Expression Profiling and Phenotypic Analysis of Downstream Targets Identify the Fox Transcription Factor Jumeau as a Master Regulator of Cardiac Progenitor Cell Division.Int J Mol Sci. 2024 Dec 1;25(23):12933. doi: 10.3390/ijms252312933. Int J Mol Sci. 2024. PMID: 39684645 Free PMC article.
-
The phospho-docking protein 14-3-3 regulates microtubule-associated proteins in oocytes including the chromosomal passenger Borealin.PLoS Genet. 2022 Jun 6;18(6):e1009995. doi: 10.1371/journal.pgen.1009995. eCollection 2022 Jun. PLoS Genet. 2022. PMID: 35666772 Free PMC article.
-
Meiosis-specific functions of kinetochore protein SPC105R required for chromosome segregation in Drosophila oocytes.Mol Biol Cell. 2024 Aug 1;35(8):ar105. doi: 10.1091/mbc.E24-02-0067. Epub 2024 Jun 12. Mol Biol Cell. 2024. PMID: 38865189 Free PMC article.
References
-
- Abad, M.A., Ruppert J.G., Buzuk L., Wear M., Zou J., Webb K.M., Kelly D.A., Voigt P., Rappsilber J., Earnshaw W.C., and Jeyaprakash A.A.. 2019. Borealin-nucleosome interaction secures chromosome association of the chromosomal passenger complex. J. Cell Biol. 218:3912–3925. 10.1083/jcb.201905040 - DOI - PMC - PubMed
-
- Adams, R.R., Maiato H., Earnshaw W.C., and Carmena M.. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865–880. 10.1083/jcb.153.4.865 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

