Myod1 and GR coordinate myofiber-specific transcriptional enhancers
- PMID: 33836079
- PMCID: PMC8096230
- DOI: 10.1093/nar/gkab226
Myod1 and GR coordinate myofiber-specific transcriptional enhancers
Abstract
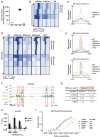
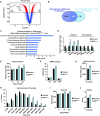
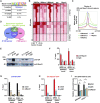
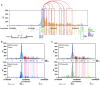
Skeletal muscle is a dynamic tissue the size of which can be remodeled through the concerted actions of various cues. Here, we investigated the skeletal muscle transcriptional program and identified key tissue-specific regulatory genetic elements. Our results show that Myod1 is bound to numerous skeletal muscle enhancers in collaboration with the glucocorticoid receptor (GR) to control gene expression. Remarkably, transcriptional activation controlled by these factors occurs through direct contacts with the promoter region of target genes, via the CpG-bound transcription factor Nrf1, and the formation of Ctcf-anchored chromatin loops, in a myofiber-specific manner. Moreover, we demonstrate that GR negatively controls muscle mass and strength in mice by down-regulating anabolic pathways. Taken together, our data establish Myod1, GR and Nrf1 as key players of muscle-specific enhancer-promoter communication that orchestrate myofiber size regulation.
© The Author(s) 2021. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Bonev B., Cavalli G.. Organization and function of the 3D genome. Nat. Rev. Genet. 2016; 17:772. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

