Targeted degradation of transcription factors by TRAFTACs: TRAnscription Factor TArgeting Chimeras
- PMID: 33836141
- PMCID: PMC8524358
- DOI: 10.1016/j.chembiol.2021.03.011
Targeted degradation of transcription factors by TRAFTACs: TRAnscription Factor TArgeting Chimeras
Abstract
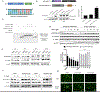
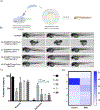
Many diseases, including cancer, stem from aberrant activation or overexpression of oncoproteins that are associated with multiple signaling pathways. Although proteins with catalytic activity can be successfully drugged, the majority of other protein families, such as transcription factors, remain intractable due to their lack of ligandable sites. In this study, we report the development of TRAnscription Factor TArgeting Chimeras (TRAFTACs) as a generalizable strategy for targeted transcription factor degradation. We show that TRAFTACs, which consist of a chimeric oligonucleotide that simultaneously binds to the transcription factor of interest (TOI) and to HaloTag-fused dCas9 protein, can induce degradation of the former via the proteasomal pathway. Application of TRAFTACs to two oncogenic TOIs, NF-κB and brachyury, suggests that TRAFTACs can be successfully employed for the targeted degradation of other DNA-binding proteins. Thus, TRAFTAC technology is potentially a generalizable strategy to induce degradation of other transcription factors both in vitro and in vivo.
Keywords: E3 ligase; HaloTag; PROTACs; brachyury; dCas9; degradation; proteasome; transcription factors; undruggable; zebrafish.
Published by Elsevier Ltd.
Conflict of interest statement
Declaration of interests C.M.C. is founder, shareholder, and consultant to Arvinas, Inc. and Halda, LLC, which support research in his laboratory.
Figures



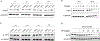

Comment in
-
Taming transcription factors with TRAFTACs.Cell Chem Biol. 2021 May 20;28(5):588-590. doi: 10.1016/j.chembiol.2021.04.016. Cell Chem Biol. 2021. PMID: 34019844
Similar articles
-
OligoTRAFTACs: A generalizable method for transcription factor degradation.RSC Chem Biol. 2022 Jul 26;3(9):1144-1153. doi: 10.1039/d2cb00138a. eCollection 2022 Aug 31. RSC Chem Biol. 2022. PMID: 36128504 Free PMC article.
-
Multifaceted targeted protein degradation systems for different cellular compartments.Bioessays. 2022 Jun;44(6):e2200008. doi: 10.1002/bies.202200008. Epub 2022 Apr 13. Bioessays. 2022. PMID: 35417040 Review.
-
PROTACs: An Emerging Targeting Technique for Protein Degradation in Drug Discovery.Bioessays. 2018 Apr;40(4):e1700247. doi: 10.1002/bies.201700247. Epub 2018 Feb 23. Bioessays. 2018. PMID: 29473971 Review.
-
Targeted Protein Degradation by Chimeric Small Molecules, PROTACs and SNIPERs.Front Chem. 2019 Dec 10;7:849. doi: 10.3389/fchem.2019.00849. eCollection 2019. Front Chem. 2019. PMID: 31921772 Free PMC article. Review.
-
Proteolysis targeting chimeras (PROTACs) in cancer therapy.J Exp Clin Cancer Res. 2020 Sep 15;39(1):189. doi: 10.1186/s13046-020-01672-1. J Exp Clin Cancer Res. 2020. PMID: 32933565 Free PMC article. Review.
Cited by
-
Intrinsic transcriptional heterogeneity in neuroblastoma guides mechanistic and therapeutic insights.Cell Rep Med. 2022 May 17;3(5):100632. doi: 10.1016/j.xcrm.2022.100632. Cell Rep Med. 2022. PMID: 35584622 Free PMC article. Review.
-
Recent Developments in PROTAC-Mediated Protein Degradation: From Bench to Clinic.Chembiochem. 2022 Jan 19;23(2):e202100270. doi: 10.1002/cbic.202100270. Epub 2021 Sep 23. Chembiochem. 2022. PMID: 34494353 Free PMC article. Review.
-
Expression-Driven Genetic Dependency Reveals Targets for Precision Medicine.bioRxiv [Preprint]. 2024 Oct 21:2024.10.17.618926. doi: 10.1101/2024.10.17.618926. bioRxiv. 2024. PMID: 39484404 Free PMC article. Preprint.
-
Delivering on the promise of protein degraders.Nat Rev Drug Discov. 2023 May;22(5):410-427. doi: 10.1038/s41573-023-00652-2. Epub 2023 Feb 21. Nat Rev Drug Discov. 2023. PMID: 36810917 Review.
-
Evolution of nanobodies specific for BCL11A.Proc Natl Acad Sci U S A. 2023 Jan 17;120(3):e2218959120. doi: 10.1073/pnas.2218959120. Epub 2023 Jan 10. Proc Natl Acad Sci U S A. 2023. PMID: 36626555 Free PMC article.
References
-
- BAI L, ZHOU H, XU R, ZHAO Y, CHINNASWAMY K, MCEACHERN D, CHEN J, YANG C-Y, LIU Z, WANG M, LIU L, JIANG H, WEN B, KUMAR P, MEAGHER JL, SUN D, STUCKEY JA & WANG S. 2019. A Potent and Selective Small-Molecule Degrader of STAT3 Achieves Complete Tumor Regression In Vivo. Cancer Cell, 36, 498–511.e17. - PMC - PubMed
-
- BOSOTTI R, MAGNAGHI P, DI BELLA S, COZZI L, CUSI C, BOZZI F, BELTRAMI N, CARAPEZZA G, BALLINARI D, AMBOLDI N, LUPI R, SOMASCHINI A, RADDRIZZANI L, SALOM B, GALVANI A, STACCHIOTTI S, TAMBORINI E. & ISACCHI A. 2017. Establishment and genomic characterization of the new chordoma cell line Chor-IN-1. Scientific reports, 7, 9226–9226. - PMC - PubMed
-
- BUHIMSCHI AD, ARMSTRONG HA, TOURE M, JAIME-FIGUEROA S, CHEN TL, LEHMAN AM, WOYACH JA, JOHNSON AJ, BYRD JC & CREWS CM 2018. Targeting the C481S Ibrutinib-Resistance Mutation in Bruton’s Tyrosine Kinase Using PROTAC-Mediated Degradation. Biochemistry, 57, 3564–3575. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

