Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors
- PMID: 33836143
- PMCID: PMC8185376
- DOI: 10.1016/j.stemcr.2021.03.010
Efficient conversion of human induced pluripotent stem cells into microglia by defined transcription factors
Abstract
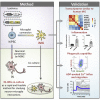
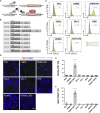
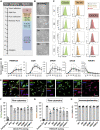
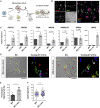
Microglia, the immune cells of the central nervous system, play critical roles in brain physiology and pathology. We report a novel approach that produces, within 10 days, the differentiation of human induced pluripotent stem cells (hiPSCs) into microglia (iMG) by forced expression of both SPI1 and CEBPA. High-level expression of the main microglial markers and the purity of the iMG cells were confirmed by RT-qPCR, immunostaining, and flow cytometry analyses. Whole-transcriptome analysis demonstrated that these iMGs resemble human fetal/adult microglia but not human monocytes. Moreover, these iMGs exhibited appropriate physiological functions, including various inflammatory responses, ADP/ATP-evoked migration, and phagocytic ability. When co-cultured with hiPSC-derived neurons, the iMGs respond and migrate toward injured neurons. This study has established a protocol for the rapid conversion of hiPSCs into functional iMGs, which should facilitate functional studies of human microglia using different disease models and also help with drug discovery.
Keywords: CEBPA; SPI1/PU.1; induced microglia; induced pluripotent stem cells; reprogramming.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Buttgereit A., Lelios I., Yu X., Vrohlings M., Krakoski N.R., Gautier E.L., Nishinakamura R., Becher B., Greter M. Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 2016;17:1397–1406. - PubMed
-
- Caiazzo M., Dell'Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

