Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant
- PMID: 33836314
- PMCID: PMC8011323
- DOI: 10.1016/j.jcv.2021.104814
Implementation of an in-house real-time reverse transcription-PCR assay for the rapid detection of the SARS-CoV-2 Marseille-4 variant
Abstract
Introduction: The SARS-CoV-2 pandemic has been associated with the occurrence since summer 2020 of several viral variants that overlapped or succeeded each other in time. Those of current concern harbor mutations within the spike receptor binding domain (RBD) that may be associated with viral escape to immune responses. In our geographical area a viral variant we named Marseille-4 harbors a S477 N substitution in this RBD.
Materials and methods: We aimed to implement an in-house one-step real-time reverse transcription-PCR (qPCR) assay with a hydrolysis probe that specifically detects the SARS-CoV-2 Marseille-4 variant.
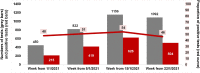
Results: All 6 cDNA samples from Marseille-4 variant strains identified in our institute by genome next-generation sequencing (NGS) tested positive using our Marseille-4 specific qPCR, whereas all 32 cDNA samples from other variants tested negative. In addition, 39/42 (93 %) respiratory samples identified by NGS as containing a Marseille-4 variant strain and 0/26 samples identified as containing non-Marseille-4 variant strains were positive. Finally, 2018/3960 (51%) patients SARS-CoV-2-diagnosed in our institute, 10/277 (3.6 %) respiratory samples collected in Algeria, and none of 207 respiratory samples collected in Senegal, Morocco, or Lebanon tested positive using our Marseille-4 specific qPCR.
Discussion: Our in-house qPCR system was found reliable to detect specifically the Marseille-4 variant and allowed estimating it is involved in about half of our SARS-CoV-2 diagnoses since December 2020. Such approach allows the real-time surveillance of SARS-CoV-2 variants, which is warranted to monitor and assess their epidemiological and clinical characterics based on comprehensive sets of data.
Keywords: Covid-19; Diagnosis; Marseille-4; Molecular epidemiology; SARS-CoV-2; Variant; qPCR.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors report no declarations of interest.
Figures

References
-
- Tegally H., Wilkinson E., Giovanetti M., et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;9(March) doi: 10.1038/s41586-021-03402-9. Epub ahead of print. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

