Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns
- PMID: 33836835
- PMCID: PMC8033687
- DOI: 10.1186/s40164-021-00207-4
Profiling of somatic mutations and fusion genes in acute myeloid leukemia patients with FLT3-ITD or FLT3-TKD mutation at diagnosis reveals distinct evolutionary patterns
Abstract
Background: The receptor tyrosine kinase FLT3 with internal tandem duplications within the juxtamembrane domain (FLT3-ITD) is a poor prognostic factor; however, the prognostic significance of missense mutation in the tyrosine kinase domain (FLT3-TKD) is controversial. Furthermore, the accompanying mutations and fusion genes with FLT3 mutations are unclear in acute myeloid leukemia (AML).
Methods: We investigated FLT3 mutations and their correlation with other gene mutations and gene fusions through two RNA-seq based next-generation sequencing (NGS) method and prognostic impact in 207 de novo AML patients.
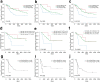
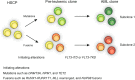
Results: FLT3-ITD mutations were positive in 58 patients (28%), and FLT3-TKD mutations were positive in 20 patients (9.7%). FLT3-ITD was associated with a higher white blood cell count (WBC, mean 72.9 × 109/L vs. 24.2 × 109/L, P = 0.000), higher bone marrow blasts (mean 65.9% vs. 56.0%, P = 0.024), and NK-AML (normal karyotype) (64.8% vs. 48.4%, P = 0.043). NPM1 and DNMT3A mutations were enriched in FLT3-ITD (53.5% vs. 15.3%, P = 0.000; 34.6% vs. 13%, P = 0.003). However, the mutations of CEBPA were excluded in FLT3-AML (3.8% vs. 0% vs. 19.8%, P = 0.005). Mutations of Ras and TP53 were unlikely associated with FLT3-ITD (1.9% vs. 20.6%, P = 0.006; 0% vs. 6.1%, P = 0.04). The common fusion genes (> 10%) in FLT3-ITD had MLL-rearrangement and NUP98-rearrangement, while the common fusion genes in FLT3-TKD had AML1-ETO and MLL-rearrangement. Two novel fusion genes PRDM16-SKI and EFAN2-ZNF238 were identified in FLT3-ITD patients. Gene fusions and NPM1 mutation were mutually excluded in FLT3-ITD and FLT3-TKD patients. Their patterns of mutual exclusivity and cooperation among mutated genes suggest that additional driver genetic alterations are required and reveal two evolutionary patterns of FLT3 pathogenesis. Patients with FLT3-ITD had a lower CR (complete remission) rate, lower 3-year OS (overall survival), DFS (disease-free survival), and EFS (event-free survival) compared to FLT3wtAML. NK-AML with FLT3-ITD had a lower 3-year OS, DFS, and EFS than those without, while FLT3-TKD did not influence the survival in whole cohort and NK-AML. Besides, we found that FLT3-ITD/TET2 bimutation defined a poor prognostic subgroup.
Conclusions: Our study offers deep insights into the molecular pathogenesis and biology of AML with FLT3-ITD and FLT3-TKD by providing the profiles of concurrent molecular alterations and the clinical impact of FLT3-ITD and FLT3-TKD on AML patients.
Keywords: Acute myeloid leukemia; FLT3-ITD; FLT3-TKD; Next-generation sequencing; TET2.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
[Clinical Characteristics and Prognostic Relevance of Co-Mutated Genes in Acute Myeloid Leukemia Patients with FLT3 Mutations].Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2024 Aug;32(4):1032-1038. doi: 10.19746/j.cnki.issn.1009-2137.2024.04.009. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2024. PMID: 39192394 Chinese.
-
FLT3-TKD mutation in childhood acute myeloid leukemia.Leukemia. 2003 May;17(5):883-6. doi: 10.1038/sj.leu.2402928. Leukemia. 2003. PMID: 12750701
-
[Prevalence and clinical significance of FLT3 mutations in acute promyelocytic leukemia].Zhonghua Xue Ye Xue Za Zhi. 2008 Nov;29(11):757-61. Zhonghua Xue Ye Xue Za Zhi. 2008. PMID: 19176014 Chinese.
-
FLT3-TKD in the prognosis of patients with acute myeloid leukemia: A meta-analysis.Front Oncol. 2023 Feb 17;13:1086846. doi: 10.3389/fonc.2023.1086846. eCollection 2023. Front Oncol. 2023. PMID: 36874106 Free PMC article.
-
[FLT3 Mutations in Acute Myeloid Leukemia].Rinsho Byori. 2017 Jan;65(1):44-51. Rinsho Byori. 2017. PMID: 30695511 Review. Japanese.
Cited by
-
Recent Advances in the Development of Anti-FLT3 CAR T-Cell Therapies for Treatment of AML.Biomedicines. 2022 Sep 30;10(10):2441. doi: 10.3390/biomedicines10102441. Biomedicines. 2022. PMID: 36289703 Free PMC article. Review.
-
Dynamin2 mutations in newly diagnosed acute myeloid leukemia: clinical characteristics, and prognostic significance.Exp Hematol Oncol. 2025 Mar 21;14(1):42. doi: 10.1186/s40164-025-00628-5. Exp Hematol Oncol. 2025. PMID: 40119466 Free PMC article.
-
Clinical analysis of 82 cases of acute promyelocytic leukemia with PML-RARα short isoform in children and adults.Front Oncol. 2024 Feb 21;14:1342671. doi: 10.3389/fonc.2024.1342671. eCollection 2024. Front Oncol. 2024. PMID: 38450185 Free PMC article.
-
Integrated analysis and validation reveal CYTH4 as a potential prognostic biomarker in acute myeloid leukemia.Oncol Lett. 2024 Jan 16;27(3):103. doi: 10.3892/ol.2024.14236. eCollection 2024 Mar. Oncol Lett. 2024. PMID: 38298432 Free PMC article.
-
Targeting Proliferation Signals and the Cell Cycle Machinery in Acute Leukemias: Novel Molecules on the Horizon.Molecules. 2023 Jan 26;28(3):1224. doi: 10.3390/molecules28031224. Molecules. 2023. PMID: 36770891 Free PMC article. Review.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

