Creating enzymes and self-sufficient cells for biosynthesis of the non-natural cofactor nicotinamide cytosine dinucleotide
- PMID: 33837188
- PMCID: PMC8035330
- DOI: 10.1038/s41467-021-22357-z
Creating enzymes and self-sufficient cells for biosynthesis of the non-natural cofactor nicotinamide cytosine dinucleotide
Abstract
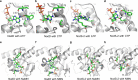
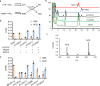
Nicotinamide adenine dinucleotide (NAD) and its reduced form are indispensable cofactors in life. Diverse NAD mimics have been developed for applications in chemical and biological sciences. Nicotinamide cytosine dinucleotide (NCD) has emerged as a non-natural cofactor to mediate redox transformations, while cells are fed with chemically synthesized NCD. Here, we create NCD synthetase (NcdS) by reprograming the substrate binding pockets of nicotinic acid mononucleotide (NaMN) adenylyltransferase to favor cytidine triphosphate and nicotinamide mononucleotide over their regular substrates ATP and NaMN, respectively. Overexpression of NcdS alone in the model host Escherichia coli facilitated intracellular production of NCD, and higher NCD levels up to 5.0 mM were achieved upon further pathway regulation. Finally, the non-natural cofactor self-sufficiency was confirmed by mediating an NCD-linked metabolic circuit to convert L-malate into D-lactate. NcdS together with NCD-linked enzymes offer unique tools and opportunities for intriguing studies in chemical biology and synthetic biology.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Engineering Escherichia coli Nicotinic Acid Mononucleotide Adenylyltransferase for Fully Active Amidated NAD Biosynthesis.Appl Environ Microbiol. 2017 Jun 16;83(13):e00692-17. doi: 10.1128/AEM.00692-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28455340 Free PMC article.
-
Characterization and application of a novel nicotinamide mononucleotide adenylyltransferase from Thermus thermophilus HB8.J Biosci Bioeng. 2018 Apr;125(4):385-389. doi: 10.1016/j.jbiosc.2017.10.017. Epub 2017 Nov 27. J Biosci Bioeng. 2018. PMID: 29175123
-
Creation of bioorthogonal redox systems depending on nicotinamide flucytosine dinucleotide.J Am Chem Soc. 2011 Dec 28;133(51):20857-62. doi: 10.1021/ja2074032. Epub 2011 Dec 6. J Am Chem Soc. 2011. PMID: 22098020
-
Structure and function of nicotinamide mononucleotide adenylyltransferase.Curr Med Chem. 2004 Apr;11(7):873-85. doi: 10.2174/0929867043455666. Curr Med Chem. 2004. PMID: 15078171 Review.
-
The NMN/NaMN adenylyltransferase (NMNAT) protein family.Front Biosci (Landmark Ed). 2009 Jan 1;14(2):410-31. doi: 10.2741/3252. Front Biosci (Landmark Ed). 2009. PMID: 19273075 Review.
Cited by
-
Engineering a newly identified alcohol dehydrogenase from Sphingobium Sp. for efficient utilization of nicotinamide cofactors biomimetics.Bioresour Bioprocess. 2025 May 5;12(1):41. doi: 10.1186/s40643-025-00870-z. Bioresour Bioprocess. 2025. PMID: 40325297 Free PMC article.
-
Shifting redox reaction equilibria on demand using an orthogonal redox cofactor.Nat Chem Biol. 2024 Nov;20(11):1535-1546. doi: 10.1038/s41589-024-01702-5. Epub 2024 Aug 13. Nat Chem Biol. 2024. PMID: 39138383 Free PMC article.
-
A phosphite-based screening platform for identification of enzymes favoring nonnatural cofactors.Sci Rep. 2022 Jul 21;12(1):12484. doi: 10.1038/s41598-022-16599-0. Sci Rep. 2022. PMID: 35864126 Free PMC article.
-
Artificial Small Molecules as Cofactors and Biomacromolecular Building Blocks in Synthetic Biology: Design, Synthesis, Applications, and Challenges.Molecules. 2023 Aug 3;28(15):5850. doi: 10.3390/molecules28155850. Molecules. 2023. PMID: 37570818 Free PMC article. Review.
-
Truncating the C terminus of formate dehydrogenase leads to improved preference to nicotinamide cytosine dinucleotide.Sci Rep. 2024 Nov 20;14(1):28701. doi: 10.1038/s41598-024-79885-z. Sci Rep. 2024. PMID: 39562703 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

