Bark-dwelling methanotrophic bacteria decrease methane emissions from trees
- PMID: 33837213
- PMCID: PMC8035153
- DOI: 10.1038/s41467-021-22333-7
Bark-dwelling methanotrophic bacteria decrease methane emissions from trees
Abstract
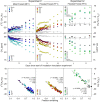
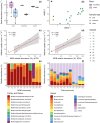
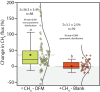
Tree stems are an important and unconstrained source of methane, yet it is uncertain whether internal microbial controls (i.e. methanotrophy) within tree bark may reduce methane emissions. Here we demonstrate that unique microbial communities dominated by methane-oxidising bacteria (MOB) dwell within bark of Melaleuca quinquenervia, a common, invasive and globally distributed lowland species. In laboratory incubations, methane-inoculated M. quinquenervia bark mediated methane consumption (up to 96.3 µmol m-2 bark d-1) and reveal distinct isotopic δ13C-CH4 enrichment characteristic of MOB. Molecular analysis indicates unique microbial communities reside within the bark, with MOB primarily from the genus Methylomonas comprising up to 25 % of the total microbial community. Methanotroph abundance was linearly correlated to methane uptake rates (R2 = 0.76, p = 0.006). Finally, field-based methane oxidation inhibition experiments demonstrate that bark-dwelling MOB reduce methane emissions by 36 ± 5 %. These multiple complementary lines of evidence indicate that bark-dwelling MOB represent a potentially significant methane sink, and an important frontier for further research.
Conflict of interest statement
The authors declare no competing interests
Figures
References
-
- Neubauer SC, Megonigal JP. Moving beyond global warming potentials to quantify the climatic role of ecosystems. Ecosystems. 2015;18:1000–1013. doi: 10.1007/s10021-015-9879-4. - DOI
-
- Saunois, M. et al. The global methane budget 2000-2017. Earth Syst. Sci. Data10.5194/essd-2019-128 (2019).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

