Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity
- PMID: 33837217
- PMCID: PMC8035321
- DOI: 10.1038/s41467-021-22397-5
Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity
Abstract
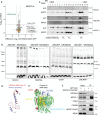
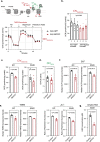
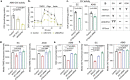
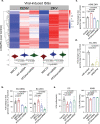
Mito-SEPs are small open reading frame-encoded peptides that localize to the mitochondria to regulate metabolism. Motivated by an intriguing negative association between mito-SEPs and inflammation, here we screen for mito-SEPs that modify inflammatory outcomes and report a mito-SEP named "Modulator of cytochrome C oxidase during Inflammation" (MOCCI) that is upregulated during inflammation and infection to promote host-protective resolution. MOCCI, a paralog of the NDUFA4 subunit of cytochrome C oxidase (Complex IV), replaces NDUFA4 in Complex IV during inflammation to lower mitochondrial membrane potential and reduce ROS production, leading to cyto-protection and dampened immune response. The MOCCI transcript also generates miR-147b, which targets the NDUFA4 mRNA with similar immune dampening effects as MOCCI, but simultaneously enhances RIG-I/MDA-5-mediated viral immunity. Our work uncovers a dual-component pleiotropic regulation of host inflammation and immunity by MOCCI (C15ORF48) for safeguarding the host during infection and inflammation.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

