Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes
- PMID: 33837218
- PMCID: PMC8035407
- DOI: 10.1038/s41467-021-22331-9
Single-cell RNA sequencing reveals the mesangial identity and species diversity of glomerular cell transcriptomes
Abstract
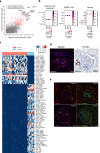
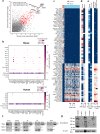
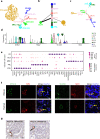
Molecular characterization of the individual cell types in human kidney as well as model organisms are critical in defining organ function and understanding translational aspects of biomedical research. Previous studies have uncovered gene expression profiles of several kidney glomerular cell types, however, important cells, including mesangial (MCs) and glomerular parietal epithelial cells (PECs), are missing or incompletely described, and a systematic comparison between mouse and human kidney is lacking. To this end, we use Smart-seq2 to profile 4332 individual glomerulus-associated cells isolated from human living donor renal biopsies and mouse kidney. The analysis reveals genetic programs for all four glomerular cell types (podocytes, glomerular endothelial cells, MCs and PECs) as well as rare glomerulus-associated macula densa cells. Importantly, we detect heterogeneity in glomerulus-associated Pdgfrb-expressing cells, including bona fide intraglomerular MCs with the functionally active phagocytic molecular machinery, as well as a unique mural cell type located in the central stalk region of the glomerulus tuft. Furthermore, we observe remarkable species differences in the individual gene expression profiles of defined glomerular cell types that highlight translational challenges in the field and provide a guide to design translational studies.
Conflict of interest statement
AstraZeneca is financially supporting C.B., R.S. and J.P. laboratories, M.L. is an employee of AstraZeneca, C.B. is a consultant for AstraZeneca. The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

