Multinucleation associated DNA damage blocks proliferation in p53-compromised cells
- PMID: 33837239
- PMCID: PMC8035210
- DOI: 10.1038/s42003-021-01979-5
Multinucleation associated DNA damage blocks proliferation in p53-compromised cells
Abstract
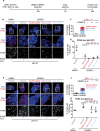
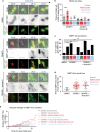
Nuclear atypia is one of the hallmarks of cancers. Here, we perform single-cell tracking studies to determine the immediate and long-term impact of nuclear atypia. Tracking the fate of newborn cells exhibiting nuclear atypia shows that multinucleation, unlike other forms of nuclear atypia, blocks proliferation in p53-compromised cells. Because ~50% of cancers display compromised p53, we explored how multinucleation blocks proliferation. Multinucleation increases 53BP1-decorated nuclear bodies (DNA damage repair platforms), along with a heterogeneous reduction in transcription and protein accumulation across the multi-nucleated compartments. Multinucleation Associated DNA Damage associated with 53BP1-bodies remains unresolved for days, despite an intact NHEJ machinery that repairs laser-induced DNA damage within minutes. Persistent DNA damage, a DNA replication block, and reduced phospho-Rb, reveal a novel replication stress independent cell cycle arrest caused by mitotic lesions. These findings call for segregating protective and prohibitive nuclear atypia to inform therapeutic approaches aimed at limiting tumour heterogeneity.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

