Intracellular mRNA transport and localized translation
- PMID: 33837370
- PMCID: PMC9346928
- DOI: 10.1038/s41580-021-00356-8
Intracellular mRNA transport and localized translation
Erratum in
-
Author Correction: Intracellular mRNA transport and localized translation.Nat Rev Mol Cell Biol. 2021 Jul;22(7):505. doi: 10.1038/s41580-021-00374-6. Nat Rev Mol Cell Biol. 2021. PMID: 33911234 No abstract available.
Abstract
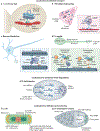
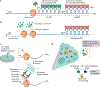
Fine-tuning cellular physiology in response to intracellular and environmental cues requires precise temporal and spatial control of gene expression. High-resolution imaging technologies to detect mRNAs and their translation state have revealed that all living organisms localize mRNAs in subcellular compartments and create translation hotspots, enabling cells to tune gene expression locally. Therefore, mRNA localization is a conserved and integral part of gene expression regulation from prokaryotic to eukaryotic cells. In this Review, we discuss the mechanisms of mRNA transport and local mRNA translation across the kingdoms of life and at organellar, subcellular and multicellular resolution. We also discuss the properties of messenger ribonucleoprotein and higher order RNA granules and how they may influence mRNA transport and local protein synthesis. Finally, we summarize the technological developments that allow us to study mRNA localization and local translation through the simultaneous detection of mRNAs and proteins in single cells, mRNA and nascent protein single-molecule imaging, and bulk RNA and protein detection methods.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures




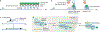
References
-
- Jeffery WR, Tomlinson CR & Brodeur RD Localization of actin messenger RNA during early ascidian development. Dev. Biol 99, 408–417 (1983). - PubMed
-
- Rebagliati MR, Weeks DL, Harvey RP & Melton DA Identification and cloning of localized maternal RNAs from Xenopus eggs. Cell 42, 769–777 (1985). - PubMed
-
- Bopp D, Burri M, Baumgartner S, Frigerio G & Noll M Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell 47, 1033–1040 (1986). - PubMed
-
- Lawrence JB & Singer RH Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45, 407–415 (1986). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

