Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation
- PMID: 33837568
- PMCID: PMC8251148
- DOI: 10.1111/all.14850
Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation
Abstract
Background: First vaccines for prevention of Coronavirus disease 2019 (COVID-19) are becoming available but there is a huge and unmet need for specific forms of treatment. In this study we aimed to evaluate the anti-SARS-CoV-2 effect of siRNA both in vitro and in vivo.
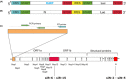
Methods: To identify the most effective molecule out of a panel of 15 in silico designed siRNAs, an in vitro screening system based on vectors expressing SARS-CoV-2 genes fused with the firefly luciferase reporter gene and SARS-CoV-2-infected cells was used. The most potent siRNA, siR-7, was modified by Locked nucleic acids (LNAs) to obtain siR-7-EM with increased stability and was formulated with the peptide dendrimer KK-46 for enhancing cellular uptake to allow topical application by inhalation of the final formulation - siR-7-EM/KK-46. Using the Syrian Hamster model for SARS-CoV-2 infection the antiviral capacity of siR-7-EM/KK-46 complex was evaluated.
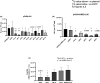
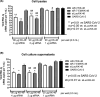
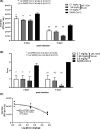
Results: We identified the siRNA, siR-7, targeting SARS-CoV-2 RNA-dependent RNA polymerase (RdRp) as the most efficient siRNA inhibiting viral replication in vitro. Moreover, we showed that LNA-modification and complexation with the designed peptide dendrimer enhanced the antiviral capacity of siR-7 in vitro. We demonstrated significant reduction of virus titer and lung inflammation in animals exposed to inhalation of siR-7-EM/KK-46 in vivo.
Conclusions: Thus, we developed a therapeutic strategy for COVID-19 based on inhalation of a modified siRNA-peptide dendrimer formulation. The developed medication is intended for inhalation treatment of COVID-19 patients.
Keywords: COVID-19; LNA; SARS-CoV-2; peptide dendrimers; siRNA.
© 2021 EAACI and John Wiley and Sons A/S. Published by John Wiley and Sons Ltd.
Conflict of interest statement
Rudolf Valenta has received research grants from the Austrian Science Fund (FWF), HVD Biotech, Vienna, Austria, Worg Pharmaceuticals, Hangzhou, China and Viravaxx, Vienna, Austria and serves as a consultant for Viravaxx. Veronica Skvortsova currently serves as head of the Federal Medico‐biological Agency of Russia (FMBA Russia). Musa Khaitov, Alexandra Nikonova, Ksenia Kozhikova, Ilya Kofiadi, Igor Shilovskiy, Valeriy Smirnov, Ivan Kozlov, Sergey Andreev, Olesya Koloskova and Ilya Sergeev are authors on a patent application related to this study. The other authors do not have any conflicts of interest to declare.
Figures


References
-
- Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS‐CoV‐2 in Wuhan, China. Allergy 2020;75(7):1730‐1741. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

