CDR1as regulated by hnRNPM maintains stemness of periodontal ligament stem cells via miR-7/KLF4
- PMID: 33837664
- PMCID: PMC8093972
- DOI: 10.1111/jcmm.16541
CDR1as regulated by hnRNPM maintains stemness of periodontal ligament stem cells via miR-7/KLF4
Abstract
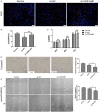
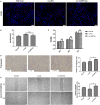
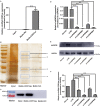
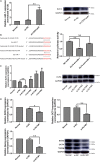
CDR1as is a well-identified circular RNA with regulatory roles in a variety of physiological processes. However, the effects of CDR1as on stemness of periodontal ligament stem cells (PDLSCs) and the underlying mechanisms remain unclear. In this study, we detect CDR1as in human PDLSCs, and subsequently demonstrate that CDR1as maintains PDLSC stemness. Knockdown of CDR1as decreases the expression levels of stemness-related genes and impairs the cell's multi-differentiation and cell migration abilities, while overexpression of CDR1as increases the expression levels of stemness-related genes and enhances these abilities. Furthermore, our results indicate that the RNA-binding protein hnRNPM directly interacts with CDR1as and regulates its expression in PDLSCs. In addition, we show that CDR1as promotes the expression of stemness-related genes in PDLSCs by inhibiting miR-7-mediated suppression of KLF4 expression. Collectively, our results demonstrate that CDR1as participates in the molecular circuitry that regulates PDLSC stemness.
Keywords: KLF4; circRNA CDR1as; hnRNPM; miR-7; periodontal ligament stem cell; stemness.
© 2021 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway.Stem Cell Res Ther. 2018 Aug 31;9(1):232. doi: 10.1186/s13287-018-0976-0. Stem Cell Res Ther. 2018. PMID: 30170617 Free PMC article.
-
circRNA CDR1as Regulated the Proliferation of Human Periodontal Ligament Stem Cells under a Lipopolysaccharide-Induced Inflammatory Condition.Mediators Inflamm. 2019 Sep 8;2019:1625381. doi: 10.1155/2019/1625381. eCollection 2019. Mediators Inflamm. 2019. PMID: 31582895 Free PMC article.
-
Restoration of miR-1305 relieves the inhibitory effect of nicotine on periodontal ligament-derived stem cell proliferation, migration, and osteogenic differentiation.J Oral Pathol Med. 2017 Apr;46(4):313-320. doi: 10.1111/jop.12492. Epub 2016 Sep 8. J Oral Pathol Med. 2017. PMID: 27604968
-
TAZ promotes the proliferation and osteogenic differentiation of human periodontal ligament stem cells via the p-SMAD3.J Cell Biochem. 2020 Feb;121(2):1101-1113. doi: 10.1002/jcb.29346. Epub 2019 Sep 3. J Cell Biochem. 2020. PMID: 31478222
-
The role of CDR1as/ciRS-7 in cardio-cerebrovascular diseases.Biomed Pharmacother. 2023 Nov;167:115589. doi: 10.1016/j.biopha.2023.115589. Epub 2023 Sep 28. Biomed Pharmacother. 2023. PMID: 37776642 Review.
Cited by
-
Upregulation of circGDI2 inhibits tumorigenesis by stabilizing the expression of RNA m6A demethylase FTO in oral squamous cell carcinoma.Noncoding RNA Res. 2024 Aug 9;10:140-152. doi: 10.1016/j.ncrna.2024.08.001. eCollection 2025 Feb. Noncoding RNA Res. 2024. PMID: 39399378 Free PMC article.
-
MicroRNAs Function in Dental Stem Cells as a Promising Biomarker and Therapeutic Target for Dental Diseases.Mol Diagn Ther. 2023 Nov;27(6):703-722. doi: 10.1007/s40291-023-00675-w. Epub 2023 Sep 29. Mol Diagn Ther. 2023. PMID: 37773247 Review.
-
Circular RNA-Mediated Regulation of Oral Tissue-Derived Stem Cell Differentiation: Implications for Oral Medicine and Orthodontic Applications.Stem Cell Rev Rep. 2024 Apr;20(3):656-671. doi: 10.1007/s12015-024-10683-w. Epub 2024 Jan 27. Stem Cell Rev Rep. 2024. PMID: 38279054 Free PMC article. Review.
-
The emerging roles of circRNAs in cancer and oncology.Nat Rev Clin Oncol. 2022 Mar;19(3):188-206. doi: 10.1038/s41571-021-00585-y. Epub 2021 Dec 15. Nat Rev Clin Oncol. 2022. PMID: 34912049 Review.
-
Epigenetic regulation of dental-derived stem cells and their application in pulp and periodontal regeneration.PeerJ. 2023 Jan 3;11:e14550. doi: 10.7717/peerj.14550. eCollection 2023. PeerJ. 2023. PMID: 36620748 Free PMC article. Review.
References
-
- Seo BM, Miura M, Gronthos S, et al. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149‐155. - PubMed
-
- Hu L, Liu Y, Wang S. Stem cell‐based tooth and periodontal regeneration. Oral Dis. 2018;24:696‐705. - PubMed
-
- Bright R, Hynes K, Gronthos S, Bartold PM. Periodontal ligament‐derived cells for periodontal regeneration in animal models: a systematic review. J Periodontal Res. 2015;50:160‐172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

