A distal Foxp3 enhancer enables interleukin-2 dependent thymic Treg cell lineage commitment for robust immune tolerance
- PMID: 33838102
- PMCID: PMC8317508
- DOI: 10.1016/j.immuni.2021.03.020
A distal Foxp3 enhancer enables interleukin-2 dependent thymic Treg cell lineage commitment for robust immune tolerance
Abstract
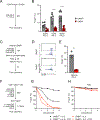
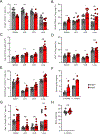
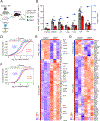
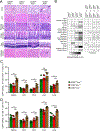
Activation of the STAT5 transcription factor downstream of the Interleukin-2 receptor (IL-2R) induces expression of Foxp3, a critical step in the differentiation of regulatory T (Treg) cells. Due to the pleiotropic effects of IL-2R signaling, it is unclear how STAT5 acts directly on the Foxp3 locus to promote its expression. Here, we report that IL-2 - STAT5 signaling converged on an enhancer (CNS0) during Foxp3 induction. CNS0 facilitated the IL-2 dependent CD25+Foxp3- precursor to Treg cell transition in the thymus. Its deficiency resulted in impaired Treg cell generation in neonates, which was partially mitigated with age. While the thymic Treg cell paucity caused by CNS0 deficiency did not result in autoimmunity on its own, it exacerbated autoimmune manifestations caused by disruption of the Aire gene. Thus, CNS0 enhancer activity ensures robust Treg cell differentiation early in postnatal life and cooperatively with other tolerance mechanisms minimizes autoimmunity.
Keywords: Foxp3; Regulatory T cells; enhancer; immune tolerance.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.Y.R. is an SAB member and a co-founder of Sonoma Biotherapeutics. All other authors declare no competing interests.
Figures



Similar articles
-
Distinct Foxp3 enhancer elements coordinate development, maintenance, and function of regulatory T cells.Immunity. 2021 May 11;54(5):947-961.e8. doi: 10.1016/j.immuni.2021.04.005. Epub 2021 Apr 29. Immunity. 2021. PMID: 33930308
-
The lineage stability and suppressive program of regulatory T cells require protein O-GlcNAcylation.Nat Commun. 2019 Jan 21;10(1):354. doi: 10.1038/s41467-019-08300-3. Nat Commun. 2019. PMID: 30664665 Free PMC article.
-
The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2.Nat Immunol. 2014 Sep;15(9):866-74. doi: 10.1038/ni.2944. Epub 2014 Jul 20. Nat Immunol. 2014. PMID: 25029551 Free PMC article.
-
T-cell tolerance and the multi-functional role of IL-2R signaling in T-regulatory cells.Immunol Rev. 2011 May;241(1):63-76. doi: 10.1111/j.1600-065X.2011.01004.x. Immunol Rev. 2011. PMID: 21488890 Free PMC article. Review.
-
Regulatory T cell identity: formation and maintenance.Trends Immunol. 2015 Jun;36(6):344-53. doi: 10.1016/j.it.2015.04.006. Epub 2015 May 13. Trends Immunol. 2015. PMID: 25981968 Free PMC article. Review.
Cited by
-
Comprehensive analysis of a novel RNA modifications-related model in the prognostic characterization, immune landscape and drug therapy of bladder cancer.Front Genet. 2023 Apr 12;14:1156095. doi: 10.3389/fgene.2023.1156095. eCollection 2023. Front Genet. 2023. PMID: 37124622 Free PMC article.
-
Foxp3 enhancers synergize to maximize regulatory T cell suppressive capacity.J Exp Med. 2021 Aug 2;218(8):e20202415. doi: 10.1084/jem.20202415. Epub 2021 Jun 4. J Exp Med. 2021. PMID: 34086055 Free PMC article.
-
Increased Frequency of Circulating Activated FOXP3+ Regulatory T Cell Subset in Patients with Chronic Lymphocytic Leukemia Is Associated with the Estimate of the Size of the Tumor Mass, STAT5 Signaling and Disease Course during Follow-Up of Patients on Therapy.Cancers (Basel). 2024 Sep 22;16(18):3228. doi: 10.3390/cancers16183228. Cancers (Basel). 2024. PMID: 39335199 Free PMC article.
-
Opposing functions of circadian protein DBP and atypical E2F family E2F8 in anti-tumor Th9 cell differentiation.Nat Commun. 2022 Oct 14;13(1):6069. doi: 10.1038/s41467-022-33733-8. Nat Commun. 2022. PMID: 36241625 Free PMC article.
-
Engineering IL-2 for immunotherapy of autoimmunity and cancer.Nat Rev Immunol. 2022 Oct;22(10):614-628. doi: 10.1038/s41577-022-00680-w. Epub 2022 Feb 25. Nat Rev Immunol. 2022. PMID: 35217787 Review.
References
-
- Anderson MS, Venanzi ES, Klein L, Chen Z, Berzins SP, Turley SJ, von Boehmer H, Bronson R, Dierich A, Benoist C, et al. (2002). Projection of an immunological self shadow within the thymus by the aire protein. Science 298, 1395–1401. - PubMed
-
- Aschenbrenner K, D’Cruz LM, Vollmann EH, Hinterberger M, Emmerich J, Swee LK, Rolink A, and Klein L (2007). Selection of Foxp3+ regulatory T cells specific for self antigen expressed and presented by Aire+ medullary thymic epithelial cells. Nat. Immunol. 8, 351–358. - PubMed
-
- Björses P, Aaltonen J, Horelli-Kuitunen N, Yaspo ML, and Peltonen L (1998). Gene defect behind APECED: a new clue to autoimmunity. Hum. Mol. Genet. 7, 1547–1553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

