The multidomain architecture of a bacteriophage endolysin enables intramolecular synergism and regulation of bacterial lysis
- PMID: 33838182
- PMCID: PMC8144678
- DOI: 10.1016/j.jbc.2021.100639
The multidomain architecture of a bacteriophage endolysin enables intramolecular synergism and regulation of bacterial lysis
Abstract
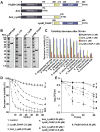
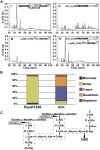
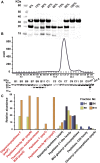
Endolysins are peptidoglycan hydrolases produced at the end of the bacteriophage (phage) replication cycle to lyse the host cell. Endolysins in Gram-positive phages come in a variety of multimodular forms that combine different catalytic and cell wall binding domains. However, the reason why phages adopt endolysins with such complex multidomain architecture is not well understood. In this study, we used the Streptococcus dysgalactiae phage endolysin PlySK1249 as a model to investigate the role of multidomain architecture in phage-induced bacterial lysis and lysis regulation. PlySK1249 consists of an amidase (Ami) domain that lyses bacterial cells, a nonbacteriolytic endopeptidase (CHAP) domain that acts as a dechaining enzyme, and a central LysM cell wall binding domain. We observed that the Ami and CHAP domains synergized for peptidoglycan digestion and bacteriolysis in the native enzyme or when expressed individually and reunified. The CHAP endopeptidase resolved complex polymers of stem-peptides to dimers and helped the Ami domain to digest peptidoglycan to completion. We also found that PlySK1249 was subject to proteolytic cleavage by host cell wall proteases both in vitro and after phage induction. Cleavage disconnected the different domains by hydrolyzing their linker regions, thus hindering their bacteriolytic cooperation and possibly modulating the lytic activity of the enzyme. PlySK1249 cleavage by cell-wall-associated proteases may represent another example of phage adaptation toward the use of existing bacterial regulation mechanism for their own advantage. In addition, understanding more thoroughly the multidomain interplay of PlySK1249 broadens our knowledge on the ideal architecture of therapeutic antibacterial endolysins.
Keywords: PlySK1249; bacteriophage; endolysin; intramolecular synergism; lysis regulation; proteolysis.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures




Similar articles
-
Potential Role of the Host-Derived Cell-Wall Binding Domain of Endolysin CD16/50L as a Molecular Anchor in Preservation of Uninfected Clostridioides difficile for New Rounds of Phage Infection.Microbiol Spectr. 2022 Apr 27;10(2):e0236121. doi: 10.1128/spectrum.02361-21. Epub 2022 Apr 4. Microbiol Spectr. 2022. PMID: 35377223 Free PMC article.
-
LambdaSa2 prophage endolysin requires Cpl-7-binding domains and amidase-5 domain for antimicrobial lysis of streptococci.FEMS Microbiol Lett. 2008 Oct;287(1):22-33. doi: 10.1111/j.1574-6968.2008.01287.x. Epub 2008 Jul 31. FEMS Microbiol Lett. 2008. PMID: 18673393
-
A novel type of peptidoglycan-binding domain highly specific for amidated D-Asp cross-bridge, identified in Lactobacillus casei bacteriophage endolysins.J Biol Chem. 2013 Jul 12;288(28):20416-26. doi: 10.1074/jbc.M112.446344. Epub 2013 Jun 3. J Biol Chem. 2013. PMID: 23733182 Free PMC article.
-
[Lysis of bacterial cells in the process of bacteriophage release--canonical and newly discovered mechanisms].Postepy Hig Med Dosw (Online). 2015 Jan 23;69:114-26. Postepy Hig Med Dosw (Online). 2015. PMID: 25614679 Review. Polish.
-
Bacteriophage endolysins as a novel class of antibacterial agents.Exp Biol Med (Maywood). 2006 Apr;231(4):366-77. doi: 10.1177/153537020623100402. Exp Biol Med (Maywood). 2006. PMID: 16565432 Review.
Cited by
-
LysSYL: a broad-spectrum phage endolysin targeting Staphylococcus species and eradicating S. aureus biofilms.Microb Cell Fact. 2024 Mar 25;23(1):89. doi: 10.1186/s12934-024-02359-4. Microb Cell Fact. 2024. PMID: 38528536 Free PMC article.
-
Identification and Characterization of a Novel Prophage Lysin against Streptococcus dysgalactiae.Molecules. 2024 Jul 20;29(14):3411. doi: 10.3390/molecules29143411. Molecules. 2024. PMID: 39064988 Free PMC article.
-
Phage endolysins are adapted to specific hosts and are evolutionarily dynamic.PLoS Biol. 2022 Aug 1;20(8):e3001740. doi: 10.1371/journal.pbio.3001740. eCollection 2022 Aug. PLoS Biol. 2022. PMID: 35913996 Free PMC article.
-
Inhibition of Listeria monocytogenes by Phage Lytic Enzymes Displayed on Tailored Bionanoparticles.Foods. 2022 Mar 17;11(6):854. doi: 10.3390/foods11060854. Foods. 2022. PMID: 35327276 Free PMC article.
-
The Synergistic and Chimeric Mechanism of Bacteriophage Endolysins: Opportunities for Application in Biotherapeutics, Food, and Health Sectors.Probiotics Antimicrob Proteins. 2025 Apr;17(2):807-831. doi: 10.1007/s12602-024-10394-1. Epub 2024 Nov 7. Probiotics Antimicrob Proteins. 2025. PMID: 39508962 Review.
References
-
- Nelson D.C., Schmelcher M., Rodriguez-Rubio L., Klumpp J., Pritchard D.G., Dong S., Donovan D.M. Endolysins as antimicrobials. Adv. Virus Res. 2012;83:299–365. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

