Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs
- PMID: 33839157
- PMCID: PMC8131913
- DOI: 10.1016/j.jbc.2021.100647
Major advances in targeted protein degradation: PROTACs, LYTACs, and MADTACs
Abstract
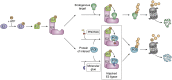
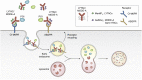
Of late, targeted protein degradation (TPD) has surfaced as a novel and innovative chemical tool and therapeutic modality. By co-opting protein degradation pathways, TPD facilitates complete removal of the protein molecules from within or outside the cell. While the pioneering Proteolysis-Targeting Chimera (PROTAC) technology and molecular glues hijack the ubiquitin-proteasome system, newer modalities co-opt autophagy or the endo-lysosomal pathway. Using this mechanism, TPD is posited to largely expand the druggable space far beyond small-molecule inhibitors. In this review, we discuss the major advances in TPD, highlight our current understanding, and explore outstanding questions in the field.
Keywords: AUTACs; LYTACs; PROTACs; chemical biology; drug action; lysosome; molecular glues; protein degradation; ubiquitination.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest C. M. C. is founder, shareholder, and consultant to Arvinas, Inc and Halda, LLC, which support research in his laboratory.
Figures


References
-
- Balch W.E., Morimoto R.I., Dillin A., Kelly J.W. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Adhikari B., Bozilovic J., Diebold M., Schwarz J.D., Hofstetter J., Schröder M., Wanior M., Narain A., Vogt M., Dudvarski Stankovic N., Baluapuri A., Schönemann L., Eing L., Bhandare P., Kuster B. PROTAC-mediated degradation reveals a non-catalytic function of AURORA-A kinase. Nat. Chem. Biol. 2020;16:1179–1188. - PMC - PubMed
-
- Powers E.T., Morimoto R.I., Dillin A., Kelly J.W., Balch W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009;78:959–991. - PubMed
-
- Dissmeyer N., Coux O., Rodriguez M.S., Barrio R. Proteostasis: A European network to break barriers and integrate science on protein homeostasis. Trends Biochem. Sci. 2019;44:383–387. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

