CRISPR-Cas systems for diagnosing infectious diseases
- PMID: 33839288
- PMCID: PMC8032595
- DOI: 10.1016/j.ymeth.2021.04.007
CRISPR-Cas systems for diagnosing infectious diseases
Abstract
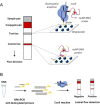
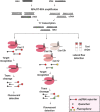
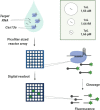
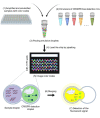
Infectious diseases are a global health problem affecting billions of people. Developing rapid and sensitive diagnostic tools is key for successful patient management and curbing disease spread. Currently available diagnostics are very specific and sensitive but time-consuming and require expensive laboratory settings and well-trained personnel; thus, they are not available in resource-limited areas, for the purposes of large-scale screenings and in case of outbreaks and epidemics. Developing new, rapid, and affordable point-of-care diagnostic assays is urgently needed. This review focuses on CRISPR-based technologies and their perspectives to become platforms for point-of-care nucleic acid detection methods and as deployable diagnostic platforms that could help to identify and curb outbreaks and emerging epidemics. We describe the mechanisms and function of different classes and types of CRISPR-Cas systems, including pros and cons for developing molecular diagnostic tests and applications of each type to detect a wide range of infectious agents. Many Cas proteins (Cas3, Cas9, Cas12, Cas13, Cas14 etc.) have been leveraged to create highly accurate and sensitive diagnostic tools combined with technologies of signal amplification and fluorescent, potentiometric, colorimetric, lateral flow assay detection and other. In particular, the most advanced platforms -- SHERLOCK/v2, DETECTR, CARMEN or CRISPR-Chip -- enable detection of attomolar amounts of pathogenic nucleic acids with specificity comparable to that of PCR but with minimal technical settings. Further developing CRISPR-based diagnostic tools promises to dramatically transform molecular diagnostics, making them easily affordable and accessible virtually anywhere in the world. The burden of socially significant diseases, frequent outbreaks, recent epidemics (MERS, SARS and the ongoing COVID-19) and outbreaks of zoonotic viruses (African Swine Fever Virus etc.) urgently need the developing and distribution of express-diagnostic tools. Recently devised CRISPR-technologies represent the unprecedented opportunity to reshape epidemiological surveillance and molecular diagnostics.
Keywords: COVID-19; HBV; HIV; HPV; Mobile phone microscopy; Molecular diagnostics; Molecular epidemiology; One pot assays; Point-of-care (POC); SARS-CoV-2; Tuberculosis; Viruses.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Gatherer D., Kohl A. Zika virus: a previously slow pandemic spreads rapidly through the Americas. J. Gen. Virol. 2016;97:269–273. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

