Trop-2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis
- PMID: 33839455
- PMCID: PMC8042651
- DOI: 10.1016/j.neo.2021.03.006
Trop-2 cleavage by ADAM10 is an activator switch for cancer growth and metastasis
Abstract
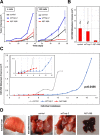
Trop-2 is a transmembrane signal transducer that can induce cancer growth. Using antibody targeting and N-terminal Edman degradation, we show here that Trop-2 undergoes cleavage in the first thyroglobulin domain loop of its extracellular region, between residues R87 and T88. Molecular modeling indicated that this cleavage induces a profound rearrangement of the Trop-2 structure, which suggested a deep impact on its biological function. No Trop-2 cleavage was detected in normal human tissues, whereas most tumors showed Trop-2 cleavage, including skin, ovary, colon, and breast cancers. Coimmunoprecipitation and mass spectrometry analysis revealed that ADAM10 physically interacts with Trop-2. Immunofluorescence/confocal time-lapse microscopy revealed that the two molecules broadly colocalize at the cell membrane. We show that ADAM10 inhibitors, siRNAs and shRNAs abolish the processing of Trop-2, which indicates that ADAM10 is an effector protease. Proteolysis of Trop-2 at R87-T88 triggered cancer cell growth both in vitro and in vivo. A corresponding role was shown for metastatic spreading of colon cancer, as the R87A-T88A Trop-2 mutant abolished xenotransplant metastatic dissemination. Activatory proteolysis of Trop-2 was recapitulated in primary human breast cancers. Together with the prognostic impact of Trop-2 and ADAM10 on cancers of the skin, ovary, colon, lung, and pancreas, these data indicate a driving role of this activatory cleavage of Trop-2 on malignant progression of tumors.
Keywords: Cell growth; Human cancer; Molecular modeling; Proteolytic processing; Signaling activation; Trop.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Ripani E, Sacchetti A, Corda D, Alberti S. The human Trop-2 is a tumor-associated calcium signal transducer. Int J Cancer. 1998;76:671–676. - PubMed
-
- Basu A, Goldenberg DM, Stein R. The epithelial/carcinoma antigen EGP-1, recognized by monoclonal antibody RS7-3G11, is phosphorylated on serine 303. Int J Cancer. 1995;62:472–479. - PubMed
-
- Klein CE, Hartmann B, Schön MP, Weber L, Alberti S. Expression of 38-kD cell-surface glycoprotein in transformed human keratinocyte cell lines, basal cell carcinomas, and epithelial germs. J Invest Dermatol. 1990;95:74–82. - PubMed
-
- Alberti S, Miotti S, Stella M, Klein CE, Fornaro M, Ménard S, Colnaghi MI. Biochemical characterization of Trop-2, a cell surface molecule expressed by human carcinomas: formal proof that the monoclonal antibodies T16 and MOv-16 recognize Trop-2. Hybridoma. 1992;11:539–545. - PubMed
-
- Calabrese G, Crescenzi C, Morizio E, Palka G, Guerra E, Alberti S. Assignment of TACSTD1 (alias TROP1, M4S1) to human chromosome 2p21 and refinement of mapping of TACSTD2 (alias TROP2, M1S1) to human chromosome 1p32 by in situ hybridization. Cytogenet Cell Genet. 2001;92:164–165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

