Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip
- PMID: 33840901
- PMCID: PMC8019248
- DOI: 10.1016/j.snb.2021.129899
Rapid point-of-care testing for SARS-CoV-2 virus nucleic acid detection by an isothermal and nonenzymatic Signal amplification system coupled with a lateral flow immunoassay strip
Abstract
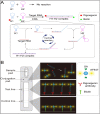
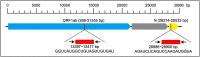
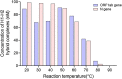
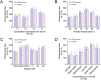
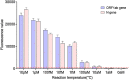
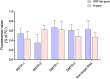
An outbreak of a new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), began in December 2019. Accurate, rapid, convenient, and relatively inexpensive diagnostic methods for SARS-CoV-2 infection are important for public health and optimal clinical care. The current gold standard for diagnosing SARS-CoV-2 infection is reverse transcription-polymerase chain reaction (RT-PCR). However, RTPCR assays are designed for use in well-equipped laboratories with sophisticated laboratory infrastructure and highly trained technicians, and are unsuitable for use in under-equipped laboratories and in the field. In this study, we report the development of an accurate, rapid, and easy-to-implement isothermal and nonenzymatic signal amplification system (a catalytic hairpin assembly (CHA) reaction) coupled with a lateral flow immunoassay (LFIA) strip-based detection method that can detect SARSCoV-2 in oropharyngeal swab samples. Our method avoids RNA isolation, PCR amplification, and elaborate result analysis, which typically takes 6-8 h. The entire CHA-LFIA detection method, from nasopharyngeal sampling to obtaining test results, takes less than 90 min. Such methods are simple and require no expensive equipment, only a simple thermostatically controlled water bath and a fluorescence reader device. We validated our method using synthetic oligonucleotides and clinical samples from 15 patients with SARS-CoV-2 infection and 15 healthy individuals. Our detection method provides a fast, simple, and sensitive (with a limit of detection (LoD) of 2000 copies/mL) alternative to the SARS-CoV-2 RT-PCR assay, with 100 % positive and negative predictive agreements.
Keywords: Catalytic hairpin assembly reaction; Lateral flow immunoassay strip; Nucleic acid test; Rapid diagnostic test; sars-cov-2.
© 2021 Published by Elsevier B.V.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

