Identification of Plitidepsin as Potent Inhibitor of SARS-CoV-2-Induced Cytopathic Effect After a Drug Repurposing Screen
- PMID: 33841165
- PMCID: PMC8033486
- DOI: 10.3389/fphar.2021.646676
Identification of Plitidepsin as Potent Inhibitor of SARS-CoV-2-Induced Cytopathic Effect After a Drug Repurposing Screen
Abstract
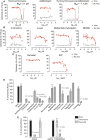
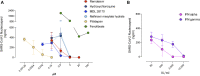
There is an urgent need to identify therapeutics for the treatment of Coronavirus disease 2019 (COVID-19). Although different antivirals are given for the clinical management of SARS-CoV-2 infection, their efficacy is still under evaluation. Here, we have screened existing drugs approved for human use in a variety of diseases, to compare how they counteract SARS-CoV-2-induced cytopathic effect and viral replication in vitro. Among the potential 72 antivirals tested herein that were previously proposed to inhibit SARS-CoV-2 infection, only 18 % had an IC50 below 25 µM or 102 IU/ml. These included plitidepsin, novel cathepsin inhibitors, nelfinavir mesylate hydrate, interferon 2-alpha, interferon-gamma, fenofibrate, camostat along the well-known remdesivir and chloroquine derivatives. Plitidepsin was the only clinically approved drug displaying nanomolar efficacy. Four of these families, including novel cathepsin inhibitors, blocked viral entry in a cell-type specific manner. Since the most effective antivirals usually combine therapies that tackle the virus at different steps of infection, we also assessed several drug combinations. Although no particular synergy was found, inhibitory combinations did not reduce their antiviral activity. Thus, these combinations could decrease the potential emergence of resistant viruses. Antivirals prioritized herein identify novel compounds and their mode of action, while independently replicating the activity of a reduced proportion of drugs which are mostly approved for clinical use. Combinations of these drugs should be tested in animal models to inform the design of fast track clinical trials.
Keywords: SARS-CoV-2; antivirals; plitidepsin; synergy; viral entry.
Copyright © 2021 Rodon, Muñoz-Basagoiti, Perez-Zsolt, Noguera-Julian, Paredes, Mateu, Quiñones, Perez, Erkizia, Blanco, Valencia, Guallar, Carrillo, Blanco, Segalés, Clotet, Vergara-Alert and Izquierdo-Useros.
Conflict of interest statement
A patent application based on this work has been filed (EP20382821.5). Unrelated to the submitted work, JB and JC are founders and shareholders of AlbaJuna Therapeutics, S.L. BC is founder and shareholder of AlbaJuna Therapeutics, S.L and AELIX Therapeutics, S.L. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

