Molecular Characterization of Enterococcus Isolates From Different Sources in Estonia Reveals Potential Transmission of Resistance Genes Among Different Reservoirs
- PMID: 33841342
- PMCID: PMC8032980
- DOI: 10.3389/fmicb.2021.601490
Molecular Characterization of Enterococcus Isolates From Different Sources in Estonia Reveals Potential Transmission of Resistance Genes Among Different Reservoirs
Abstract
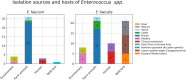
In this study, we aimed to characterize the population structure, drug resistance mechanisms, and virulence genes of Enterococcus isolates in Estonia. Sixty-one Enterococcus faecalis and 34 Enterococcus faecium isolates were collected between 2012 and 2014 across the country from various sites and sources, including farm animals and poultry (n = 53), humans (n = 12), environment (n = 24), and wild birds (n = 44). Clonal relationships of the strains were determined by whole-genome sequencing and analyzed by multi-locus sequence typing. We determined the presence of acquired antimicrobial resistance genes and 23S rRNA mutations, virulence genes, and also the plasmid or chromosomal origin of the genes using dedicated DNA sequence analysis tools available and/or homology search against an ad hoc compiled database of relevant sequences. Two E. faecalis isolates from human with vanB genes were highly resistant to vancomycin. Closely related E. faecalis strains were isolated from different host species. This indicates interspecies spread of strains and potential transfer of antibiotic resistance. Genomic context analysis of the resistance genes indicated frequent association with plasmids and mobile genetic elements. Resistance genes are often present in the identical genetic context in strains with diverse origins, suggesting the occurrence of transfer events.
Keywords: E. faecalis; E. faecium; antibiotic resistance; multi-locus sequence typing; phylogenetic analysis; van genes; virulence factors; whole-genome sequencing.
Copyright © 2021 Aun, Kisand, Laht, Telling, Kalmus, Väli, Brauer, Remm and Tenson.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Abbo L., Shukla B. S., Giles A., Aragon L., Jimenez A., Camargo J. F., et al. (2019). Linezolid-and vancomycin-resistant enterococcus faecium in solid organ transplant recipients: infection control and antimicrobial stewardship using whole genome sequencing. Clin. Infect. Dis. 69 259–265. 10.1093/cid/ciy903 - DOI - PMC - PubMed
-
- Aphis. (2014). Enterococcus on U. S. Sheep and Lamb Operations. Riverdale Park, MD: APHIS.
LinkOut - more resources
Full Text Sources
Other Literature Sources

