NAC Candidate Gene Marker for bgm-1 and Interaction With QTL for Resistance to Bean Golden Yellow Mosaic Virus in Common Bean
- PMID: 33841459
- PMCID: PMC8027503
- DOI: 10.3389/fpls.2021.628443
NAC Candidate Gene Marker for bgm-1 and Interaction With QTL for Resistance to Bean Golden Yellow Mosaic Virus in Common Bean
Abstract
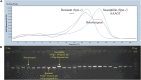
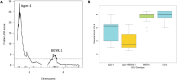
Genetic resistance is the primary means for control of Bean golden yellow mosaic virus (BGYMV) in common bean (Phaseolus vulgaris L.). Breeding for resistance is difficult because of sporadic and uneven infection across field nurseries. We sought to facilitate breeding for BGYMV resistance by improving marker-assisted selection (MAS) for the recessive bgm-1 gene and identifying and developing MAS for quantitative trait loci (QTL) conditioning resistance. Genetic linkage mapping in two recombinant inbred line populations and genome-wide association study (GWAS) in a large breeding population and two diversity panels revealed a candidate gene for bgm-1 and three QTL BGY4.1, BGY7.1, and BGY8.1 on independent chromosomes. A mutation (5 bp deletion) in a NAC (No Apical Meristem) domain transcriptional regulator superfamily protein gene Phvul.003G027100 on chromosome Pv03 corresponded with the recessive bgm-1 resistance allele. The five bp deletion in exon 2 starting at 20 bp (Pv03: 2,601,582) is expected to cause a stop codon at codon 23 (Pv03: 2,601,625), disrupting further translation of the gene. A T m -shift assay marker named PvNAC1 was developed to track bgm-1. PvNAC1 corresponded with bgm-1 across ∼1,000 lines which trace bgm-1 back to a single landrace "Garrapato" from Mexico. BGY8.1 has no effect on its own but exhibited a major effect when combined with bgm-1. BGY4.1 and BGY7.1 acted additively, and they enhanced the level of resistance when combined with bgm-1. T m -shift assay markers were generated for MAS of the QTL, but their effectiveness requires further validation.
Keywords: Phaseolus vulgaris; frameshift mutation; geminivirus; genome-wide association study; marker-assisted selection.
Copyright © 2021 Soler-Garzón, Oladzad, Beaver, Beebe, Lee, Lobaton, Macea, McClean, Raatz, Rosas, Song and Miklas.
Conflict of interest statement
The handling editor RP and the author PNM declare that they are affiliated with the INCREASE consortium on genetic resources in legumes. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
-
- Acevedo-Román M., Molina-Castañeda A., Angel-Sánchez J. C., Muñoz C. G., Beaver J. S. (2004). Inheritance of normal pod development in bean golden yellow mosaic resistant common bean. J. Am. Soc. Hortic. Sci. 129 549–552. 10.21273/JASHS.129.4.0549 - DOI
-
- Afanador L. K., Haley S. D., Kelly J. D. (1993). Adoption of a “mini-prep” DNA extraction method for RAPD marker analysis in common bean (Phaseolus vulgaris L.). Annu. Rep. Bean Improv. Coop. 36 10–11.
LinkOut - more resources
Full Text Sources
Other Literature Sources

