Genome-Based Drug Target Identification in Human Pathogen Streptococcus gallolyticus
- PMID: 33841489
- PMCID: PMC8027347
- DOI: 10.3389/fgene.2021.564056
Genome-Based Drug Target Identification in Human Pathogen Streptococcus gallolyticus
Abstract
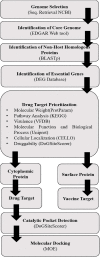
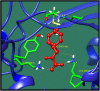
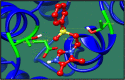
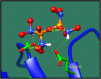
Streptococcus gallolysticus (Sg) is an opportunistic Gram-positive, non-motile bacterium, which causes infective endocarditis, an inflammation of the inner lining of the heart. As Sg has acquired resistance with the available antibiotics, therefore, there is a dire need to find new therapeutic targets and potent drugs to prevent and treat this disease. In the current study, an in silico approach is utilized to link genomic data of Sg species with its proteome to identify putative therapeutic targets. A total of 1,138 core proteins have been identified using pan genomic approach. Further, using subtractive proteomic analysis, a set of 18 proteins, essential for bacteria and non-homologous to host (human), is identified. Out of these 18 proteins, 12 cytoplasmic proteins were selected as potential drug targets. These selected proteins were subjected to molecular docking against drug-like compounds retrieved from ZINC database. Furthermore, the top docked compounds with lower binding energy were identified. In this work, we have identified novel drug and vaccine targets against Sg, of which some have already been reported and validated in other species. Owing to the experimental validation, we believe our methodology and result are significant contribution for drug/vaccine target identification against Sg-caused infective endocarditis.
Keywords: Streptococcus gallollyticus; drug prioritization; infective endocarditis; pan-genome; subtractive proteomics.
Copyright © 2021 Qureshi, Bakhtiar, Faheem, Shah, Bari, Mahmood, Sohaib, Mothana, Ullah and Jamal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


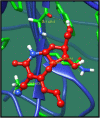
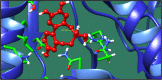
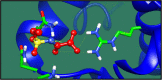
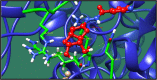
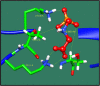
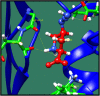
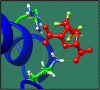

References
-
- Barh D., Jain N., Tiwari S., Parida B. P., D’Afonseca V., Li L., et al. (2011). A novel comparative genomics analysis for common drug and vaccine targets in Corynebacterium pseudotuberculosis and other CMN group of human pathogens. Chem. Biol. Drug Des. 78 73–84. 10.1111/j.1747-0285.2011.01118.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

