AutoVEM: An automated tool to real-time monitor epidemic trends and key mutations in SARS-CoV-2 evolution
- PMID: 33841748
- PMCID: PMC8020629
- DOI: 10.1016/j.csbj.2021.04.002
AutoVEM: An automated tool to real-time monitor epidemic trends and key mutations in SARS-CoV-2 evolution
Abstract
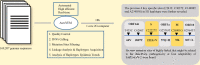
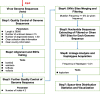
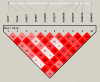
With the global epidemic of SARS-CoV-2, it is important to effectively monitor the variation, haplotype subgroup epidemic trends and key mutations of SARS-CoV-2 over time. This is of great significance to the development of new vaccines, the update of therapeutic drugs, and the improvement of detection methods. The AutoVEM tool developed in the present study could complete all mutations detections, haplotypes classification, haplotype subgroup epidemic trends and candidate key mutations analysis for 131,576 SARS-CoV-2 genome sequences in 18 h on a 1 core CPU and 2 GB RAM computer. Through haplotype subgroup epidemic trends analysis of 131,576 genome sequences, the great significance of the previous 4 specific sites (C241T, C3037T, C14408T and A23403G) was further revealed, and 6 new mutation sites of highly linked (T445C, C6286T, C22227T, G25563T, C26801G and G29645T) were discovered for the first time that might be related to the infectivity, pathogenicity or host adaptability of SARS-CoV-2. In brief, we proposed an integrative method and developed an efficient automated tool to monitor haplotype subgroup epidemic trends and screen for the candidate key mutations in the evolution of SARS-CoV-2 over time for the first time, and all data could be updated quickly to track the prevalence of previous key mutations and new candidate key mutations because of high efficiency of the tool. In addition, the idea of combinatorial analysis in the present study can also provide a reference for the mutation monitoring of other viruses.
Keywords: Automated tool; Epidemic trends; Key mutations; SARS-CoV-2.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- COVID-19. Weekly Epidemiological Update. 2020.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
