Preclinical development of a molecular clamp-stabilised subunit vaccine for severe acute respiratory syndrome coronavirus 2
- PMID: 33841880
- PMCID: PMC8021130
- DOI: 10.1002/cti2.1269
Preclinical development of a molecular clamp-stabilised subunit vaccine for severe acute respiratory syndrome coronavirus 2
Abstract
Objectives: Efforts to develop and deploy effective vaccines against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continue at pace. Here, we describe rational antigen design through to manufacturability and vaccine efficacy of a prefusion-stabilised spike (S) protein, Sclamp, in combination with the licensed adjuvant MF59 'MF59C.1' (Seqirus, Parkville, Australia).
Methods: A panel recombinant Sclamp proteins were produced in Chinese hamster ovary and screened in vitro to select a lead vaccine candidate. The structure of this antigen was determined by cryo-electron microscopy and assessed in mouse immunogenicity studies, hamster challenge studies and safety and toxicology studies in rat.
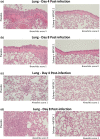
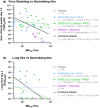
Results: In mice, the Sclamp vaccine elicits high levels of neutralising antibodies, as well as broadly reactive and polyfunctional S-specific CD4+ and cytotoxic CD8+ T cells in vivo. In the Syrian hamster challenge model (n = 70), vaccination results in reduced viral load within the lung, protection from pulmonary disease and decreased viral shedding in daily throat swabs which correlated strongly with the neutralising antibody level.
Conclusion: The SARS-CoV-2 Sclamp vaccine candidate is compatible with large-scale commercial manufacture, stable at 2-8°C. When formulated with MF59 adjuvant, it elicits neutralising antibodies and T-cell responses and provides protection in animal challenge models.
Keywords: Molecular Clamp; SARS‐CoV‐2; neutralising antibodies; polyfunctional T cells; rapid response; subunit vaccine.
© 2021 The Authors. Clinical & Translational Immunology published by John Wiley & Sons Australia, Ltd on behalf of Australian and New Zealand Society for Immunology, Inc.
Conflict of interest statement
KJC, DW and PRY are inventors of the ‘Molecular Clamp’ patent, US 2020/0040042.
Figures





References
-
- WHO(WHO) . DRAFT landscape of COVID‐19 candidate vaccines; 2021. https://www.who.int/publications/m/item/draft‐landscape‐of‐covid‐19‐cand...
-
- Chappell K, Watterson D, Young P. Chimeric Molecules and Uses Thereof US Patent 2020/0040042; 2018.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
