A proline-rich motif in the large intracellular loop of the glycine receptor α1 subunit interacts with the Pleckstrin homology domain of collybistin
- PMID: 33842008
- PMCID: PMC8020344
- DOI: 10.1016/j.jare.2020.09.009
A proline-rich motif in the large intracellular loop of the glycine receptor α1 subunit interacts with the Pleckstrin homology domain of collybistin
Abstract
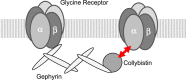
Introduction: The inhibitory glycine receptor (GlyR), a mediator of fast synaptic inhibition, is located and held at neuronal synapses through the anchoring proteins gephyrin and collybistin. Stable localization of neurotransmitter receptors is essential for synaptic function. In case of GlyRs, only beta subunits were known until now to mediate synaptic anchoring.
Objectives: We identified a poly-proline II helix (PPII) in position 365-373 of the intra-cellular TM3-4 loop of the human GlyRα1 subunit as a novel potential synaptic anchoring site. The potential role of the PPII helix as synaptic anchoring site was tested.
Methods: Glycine receptors and collybistin variants were generated and recombinantly expressed in HEK293 cells and cultured neurons. Receptor function was assessed using patch-clamp electrophysiology, protein-protein interaction was studied using co-immuno-precipitation and pulldown experiments.
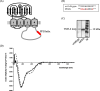
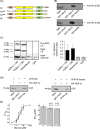
Results: Recombinantly expressed collybistin bound to isolated GlyRα1 TM3-4 loops in GST-pulldown assays. When the five proline residues P365A, P366A, P367A, P369A, P373A (GlyRα1P1-5A) located in the GlyRα1-PPII helix were replaced by alanines, the PPII secondary structure was disrupted. Recombinant GlyRα1P1-5A mutant subunits displayed normal cell surface expression and wildtype-like ion channel function, but binding to collybistin was abolished. The GlyRα1-collybistin interaction was independently confirmed by o-immunoprecipitation assays using full-length GlyRα1 subunits. Surprisingly, the interaction was not mediated by the SH3 domain of collybistin, but by its Pleckstrin homology (PH) domain. The mutation GlyRα1P366L, identified in a hyperekplexia patient, is also disrupting the PPII helix, and caused reduced collybistin binding.
Conclusion: Our data suggest a novel interaction between α1 GlyR subunits and collybistin, which is physiologically relevant in vitro and in vivo and may contribute to postsynaptic anchoring of glycine receptors.
Keywords: Collybistin; Gephyrin; Glycine receptor alpha1 subunit; Ion channel receptors; Pleckstrin homology domains; Polyproline II helix; Protein-protein interaction; SH3 domains; Synaptic anchoring.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Cairo University.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Breitinger HG. Glycine Receptors. Chichester: eLS. John Wiley & Sons Ltd; 2014.
-
- Lynch J.W. Native glycine receptor subtypes and their physiological roles. Neuropharmacology. 2009;56:303–309. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

