Machine Learning-Based Radiomics Nomogram for Detecting Extramural Venous Invasion in Rectal Cancer
- PMID: 33842316
- PMCID: PMC8033032
- DOI: 10.3389/fonc.2021.610338
Machine Learning-Based Radiomics Nomogram for Detecting Extramural Venous Invasion in Rectal Cancer
Abstract
Objective: To establish and validate a radiomics nomogram based on the features of the primary tumor for predicting preoperative pathological extramural venous invasion (EMVI) in rectal cancer using machine learning.
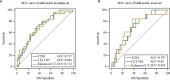
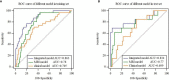
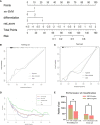
Methods: The clinical and imaging data of 281 patients with primary rectal cancer from April 2012 to May 2018 were retrospectively analyzed. All the patients were divided into a training set (n = 198) and a test set (n = 83) respectively. The radiomics features of the primary tumor were extracted from the enhanced computed tomography (CT), the T2-weighted imaging (T2WI) and the gadolinium contrast-enhanced T1-weighted imaging (CE-TIWI) of each patient. One optimal radiomics signature extracted from each modal image was generated by receiver operating characteristic (ROC) curve analysis after dimensionality reduction. Three kinds of models were constructed based on training set, including the clinical model (the optimal radiomics signature combining with the clinical features), the magnetic resonance imaging model (the optimal radiomics signature combining with the mrEMVI status) and the integrated model (the optimal radiomics signature combining with both the clinical features and the mrEMVI status). Finally, the optimal model was selected to create a radiomics nomogram. The performance of the nomogram to evaluate clinical efficacy was verified by ROC curves and decision curve analysis curves.
Results: The radiomics signature constructed based on T2WI showed the best performance, with an AUC value of 0.717, a sensitivity of 0.742 and a specificity of 0.621. The radiomics nomogram had the highest prediction efficiency, of which the AUC was 0.863, the sensitivity was 0.774 and the specificity was 0.801.
Conclusion: The radiomics nomogram had the highest efficiency in predicting EMVI. This may help patients choose the best treatment strategy and may strengthen personalized treatment methods to further optimize the treatment effect.
Keywords: computed tomography; extramural venous invasion; magnetic resonance imaging; prediction; radiomics; rectal cancer.
Copyright © 2021 Liu, Yu, Yang, Hu, Hu, Chen, Li, Zhang, Li and Lu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

