mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are Druggable Candidates for N-(2,4-Difluorophenyl)-2',4'-Difluoro-4-Hydroxybiphenyl-3-Carboxamide (NSC765598), With Consequent Anticancer Implications
- PMID: 33842373
- PMCID: PMC8034425
- DOI: 10.3389/fonc.2021.656738
mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are Druggable Candidates for N-(2,4-Difluorophenyl)-2',4'-Difluoro-4-Hydroxybiphenyl-3-Carboxamide (NSC765598), With Consequent Anticancer Implications
Abstract
Background: The application of computational and multi-omics approaches has aided our understanding of carcinogenesis and the development of therapeutic strategies. NSC765598 is a novel small molecule derivative of salicylanilide. This study aims to investigate the ligand-protein interactions of NSC765598 with its potential targets and to evaluate its anticancer activities in vitro.
Methods: We used multi-computational tools and clinical databases, respectively, to identify the potential drug target for NSC765598 and analyze the genetic profile and prognostic relevance of the targets in multiple cancers. We evaluated the in vitro anticancer activities against the National Cancer Institute 60 (NCI60) human tumor cell lines and used molecular docking to study the ligand-protein interactions. Finally, we used the DTP-COMPARE algorithm to compare the NSC765598 anticancer fingerprints with NCI standard agents.
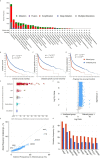
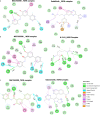
Results: We identified mammalian target of rapamycin (mTOR)/epidermal growth factor receptor (EGFR)/inducible nitric oxide synthase (iNOS)/mitogen-activated protein 2 kinase 1 (MAP2K1)/fibroblast growth factor receptor (FGFR)/transforming growth factor-β1 (TGFB1) as potential targets for NSC765598. The targets were enriched in cancer-associated pathways, were overexpressed and were of prognostic relevance in multiple cancers. Among the identified targets, genetic alterations occurred most frequently in EGFR (7%), particularly in glioblastoma, esophageal squamous cell cancer, head and neck squamous cell cancer, and non-small-cell lung cancer, and were associated with poor prognoses and survival of patients, while other targets were less frequently altered. NSC765598 displayed selective antiproliferative and cytotoxic preferences for NSCLC (50% growth inhibition (GI50) = 1.12-3.95 µM; total growth inhibition (TGI) = 3.72-16.60 μM), leukemia (GI50 = 1.20-3.10 µM; TGI = 3.90-12.70 μM), melanoma (GI50 = 1.45-3.59 µM), and renal cancer (GI50 = 1.38-3.40 µM; TGI = 4.84-13.70 μM) cell lines, while panels of colon, breast, ovarian, prostate, and central nervous system (CNS) cancer cell lines were less sensitive to NSC765598. Interestingly, NSC765598 docked well into the binding cavity of the targets by conventional H-bonds, van der Waal forces, and a variety of π-interactions, with higher preferences for EGFR (ΔG = -11.0 kcal/mol), NOS2 (ΔG = -11.0 kcal/mol), and mTOR (ΔG = -8.8 kcal/mol). NSC765598 shares similar anti-cancer fingerprints with NCI standard agents displayed acceptable physicochemical values and met the criteria of drug-likeness.
Conclusion: NSC765598 displayed significant anticancer and potential multi-target properties, thus serve as a novel candidate worthy of further preclinical studies.
Keywords: NSC765598; anticancer activity; molecular docking; multi-omics study; multi-target small molecule; protein-ligand interaction.
Copyright © 2021 Lawal, Lee, Mokgautsi, Sumitra, Khedkar, Wu and Huang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

