Oncoimmunology Meets Organs-on-Chip
- PMID: 33842539
- PMCID: PMC8032996
- DOI: 10.3389/fmolb.2021.627454
Oncoimmunology Meets Organs-on-Chip
Abstract
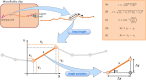
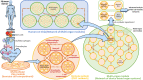
Oncoimmunology represents a biomedical research discipline coined to study the roles of immune system in cancer progression with the aim of discovering novel strategies to arm it against the malignancy. Infiltration of immune cells within the tumor microenvironment is an early event that results in the establishment of a dynamic cross-talk. Here, immune cells sense antigenic cues to mount a specific anti-tumor response while cancer cells emanate inhibitory signals to dampen it. Animals models have led to giant steps in this research context, and several tools to investigate the effect of immune infiltration in the tumor microenvironment are currently available. However, the use of animals represents a challenge due to ethical issues and long duration of experiments. Organs-on-chip are innovative tools not only to study how cells derived from different organs interact with each other, but also to investigate on the crosstalk between immune cells and different types of cancer cells. In this review, we describe the state-of-the-art of microfluidics and the impact of OOC in the field of oncoimmunology underlining the importance of this system in the advancements on the complexity of tumor microenvironment.
Keywords: Cell on Chip; Oncoimmuno chip; Organ on Chip; cancer immunology; human on chip; microfluidic device; personalized medicine; tumor microenvironment.
Copyright © 2021 Mattei, Andreone, Mencattini, De Ninno, Businaro, Martinelli and Schiavoni.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

