Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology
- PMID: 33842544
- PMCID: PMC8027086
- DOI: 10.3389/fmolb.2021.639587
Easy Synthesis of Complex Biomolecular Assemblies: Wheat Germ Cell-Free Protein Expression in Structural Biology
Abstract
Cell-free protein synthesis (CFPS) systems are gaining more importance as universal tools for basic research, applied sciences, and product development with new technologies emerging for their application. Huge progress was made in the field of synthetic biology using CFPS to develop new proteins for technical applications and therapy. Out of the available CFPS systems, wheat germ cell-free protein synthesis (WG-CFPS) merges the highest yields with the use of a eukaryotic ribosome, making it an excellent approach for the synthesis of complex eukaryotic proteins including, for example, protein complexes and membrane proteins. Separating the translation reaction from other cellular processes, CFPS offers a flexible means to adapt translation reactions to protein needs. There is a large demand for such potent, easy-to-use, rapid protein expression systems, which are optimally serving protein requirements to drive biochemical and structural biology research. We summarize here a general workflow for a wheat germ system providing examples from the literature, as well as applications used for our own studies in structural biology. With this review, we want to highlight the tremendous potential of the rapidly evolving and highly versatile CFPS systems, making them more widely used as common tools to recombinantly prepare particularly challenging recombinant eukaryotic proteins.
Keywords: NMR; cell-free protein expression; labeling; structural biology; wheat germ.
Copyright © 2021 Fogeron, Lecoq, Cole, Harbers and Böckmann.
Conflict of interest statement
MH declares competing financial interests as an employee of CellFree Sciences Co., Ltd., Japan. The other authors do not have any competing interests.
Figures



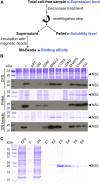

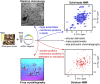

References
-
- Aceti D. J., Bingman C. A., Wrobel R. L., Frederick R. O., Makino S., Nichols K. W., et al. (2015). Expression platforms for producing eukaryotic proteins: a comparison of E. coli cell-based and wheat germ cell-free synthesis, affinity and solubility tags, and cloning strategies. J. Struct. Funct. Genomics 16, 67–80. 10.1007/s10969-015-9198-1 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

