Distinct clonal evolution of B-cells in HIV controllers with neutralizing antibody breadth
- PMID: 33843586
- PMCID: PMC8041465
- DOI: 10.7554/eLife.62648
Distinct clonal evolution of B-cells in HIV controllers with neutralizing antibody breadth
Abstract
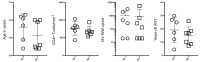
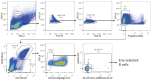
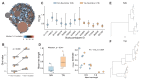
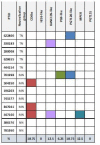
A minor subset of individuals infected with HIV-1 develop antibody neutralization breadth during the natural course of the infection, often linked to chronic, high-level viremia. Despite significant efforts, vaccination strategies have been unable to induce similar neutralization breadth and the mechanisms underlying neutralizing antibody induction remain largely elusive. Broadly neutralizing antibody responses can also be found in individuals who control HIV to low and even undetectable plasma levels in the absence of antiretroviral therapy, suggesting that high antigen exposure is not a strict requirement for neutralization breadth. We therefore performed an analysis of paired heavy and light chain B-cell receptor (BCR) repertoires in 12,591 HIV-1 envelope-specific single memory B-cells to determine alterations in the BCR immunoglobulin gene repertoire and B-cell clonal expansions that associate with neutralizing antibody breadth in 22 HIV controllers. We found that the frequency of genomic mutations in IGHV and IGLV was directly correlated with serum neutralization breadth. The repertoire of the most mutated antibodies was dominated by a small number of large clones with evolutionary signatures suggesting that these clones had reached peak affinity maturation. These data demonstrate that even in the setting of low plasma HIV antigenemia, similar to what a vaccine can potentially achieve, BCR selection for extended somatic hypermutation and clonal evolution can occur in some individuals suggesting that host-specific factors might be involved that could be targeted with future vaccine strategies.
Keywords: BCR repertoire; HIV infection; b-cell evolution; cell biology; human; infectious disease; microbiology; neutralizing antibodies.
© 2021, Cizmeci et al.
Conflict of interest statement
DC, GL, ER, AD, MS, BJ No competing interests declared, DK, GC, NN employee of Atreca Inc, YT Former employee of Atreca Inc Now employee of Esco Ventures X. GA GA is a founder of Seromyx Systems.
Figures



References
-
- CAPRISA002 Study Team. Gray ES, Madiga MC, Hermanus T, Moore PL, Wibmer CK, Tumba NL, Werner L, Mlisana K, Sibeko S, Williamson C, Abdool Karim SS, Morris L. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. Journal of Virology. 2011;85:4828–4840. doi: 10.1128/JVI.00198-11. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

