Deeper Protein Identification Using Field Asymmetric Ion Mobility Spectrometry in Top-Down Proteomics
- PMID: 33844503
- PMCID: PMC8130575
- DOI: 10.1021/acs.analchem.1c00402
Deeper Protein Identification Using Field Asymmetric Ion Mobility Spectrometry in Top-Down Proteomics
Abstract
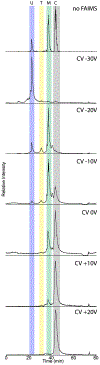
Field asymmetric ion mobility spectrometry (FAIMS), when used in proteomics studies, provides superior selectivity and enables more proteins to be identified by providing additional gas-phase separation. Here, we tested the performance of cylindrical FAIMS for the identification and characterization of proteoforms by top-down mass spectrometry of heterogeneous protein mixtures. Combining FAIMS with chromatographic separation resulted in a 62% increase in protein identifications, an 8% increase in proteoform identifications, and an improvement in proteoform identification compared to samples analyzed without FAIMS. In addition, utilization of FAIMS resulted in the identification of proteins encoded by lower-abundance mRNA transcripts. These improvements were attributable, in part, to improved signal-to-noise for proteoforms with similar retention times. Additionally, our results show that the optimal compensation voltage of any given proteoform was correlated with the molecular weight of the analyte. Collectively these results suggest that the addition of FAIMS can enhance top-down proteomics in both discovery and targeted applications.
Conflict of interest statement
The authors declare the following competing financial interest(s): N.L.K. consults for Thermo Fisher Scientific. M.B. and R.H. are employees of Thermo Fisher Scientific.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

