Two novel mouse models mimicking minor deletions in 22q11.2 deletion syndrome revealed the contribution of each deleted region to psychiatric disorders
- PMID: 33845872
- PMCID: PMC8042712
- DOI: 10.1186/s13041-021-00778-7
Two novel mouse models mimicking minor deletions in 22q11.2 deletion syndrome revealed the contribution of each deleted region to psychiatric disorders
Abstract
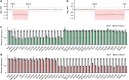
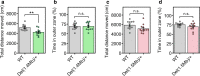
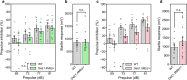
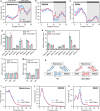
22q11.2 deletion syndrome (22q11.2DS) is a disorder caused by the segmental deletion of human chromosome 22. This chromosomal deletion is known as high genetic risk factors for various psychiatric disorders. The different deletion types are identified in 22q11.2DS patients, including the most common 3.0-Mb deletion, and the less-frequent 1.5-Mb and 1.4-Mb deletions. In previous animal studies of psychiatric disorders associated with 22q11.2DS mainly focused on the 1.5-Mb deletion and model mice mimicking the human 1.5-Mb deletion have been established with diverse genetic backgrounds, which resulted in the contradictory phenotypes. On the other hand, the contribution of the genes in 1.4-Mb region to psychiatric disorders is poorly understood. In this study, we generated two mouse lines that reproduced the 1.4-Mb and 1.5-Mb deletions of 22q11.2DS [Del(1.4 Mb)/+ and Del(1.5 Mb)/+] on the pure C57BL/6N genetic background. These mutant mice were analyzed comprehensively by behavioral tests, such as measurement of locomotor activity, sociability, prepulse inhibition and fear-conditioning memory. Del(1.4 Mb)/+ mice displayed decreased locomotor activity, but no abnormalities were observed in all other behavioral tests. Del(1.5 Mb)/+ mice showed reduction of prepulse inhibition and impairment of contextual- and cued-dependent fear memory, which is consistent with previous reports. Furthermore, apparently intact social recognition in Del(1.4 Mb)/+ and Del(1.5 Mb)/+ mice suggests that the impaired social recognition observed in Del(3.0 Mb)/+ mice mimicking the human 3.0-Mb deletion requires mutations both in 1.4-Mb and 1.5 Mb regions. Our previous study has shown that Del(3.0 Mb)/+ mice presented disturbance of behavioral circadian rhythm. Therefore, we further evaluated sleep/wakefulness cycles in Del(3.0 Mb)/+ mice by electroencephalogram (EEG) and electromyogram (EMG) recording. EEG/EMG analysis revealed the disturbed wakefulness and non-rapid eye moving sleep (NREMS) cycles in Del(3.0 Mb)/+ mice, suggesting that Del(3.0 Mb)/+ mice may be unable to maintain their wakefulness. Together, our mouse models deepen our understanding of genetic contributions to schizophrenic phenotypes related to 22q11.2DS.
Keywords: 22q11.2 deletion syndrome; Behavioral analysis; Mouse models; Rare deletion types; Sleep analysis.
Conflict of interest statement
M.K. is currently employed by Eisai Co. Ltd. The remaining authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Comprehensive analysis of a novel mouse model of the 22q11.2 deletion syndrome: a model with the most common 3.0-Mb deletion at the human 22q11.2 locus.Transl Psychiatry. 2020 Feb 5;10(1):35. doi: 10.1038/s41398-020-0723-z. Transl Psychiatry. 2020. PMID: 32066675 Free PMC article.
-
Mice lacking the circadian modulators SHARP1 and SHARP2 display altered sleep and mixed state endophenotypes of psychiatric disorders.PLoS One. 2014 Oct 23;9(10):e110310. doi: 10.1371/journal.pone.0110310. eCollection 2014. PLoS One. 2014. PMID: 25340473 Free PMC article.
-
Schizophrenia-like neurophysiological abnormalities in 22q11.2 deletion syndrome and their association to COMT and PRODH genotypes.J Psychiatr Res. 2013 Nov;47(11):1623-9. doi: 10.1016/j.jpsychires.2013.07.004. Epub 2013 Aug 1. J Psychiatr Res. 2013. PMID: 23910792
-
[Neurocognitive and psychiatric management of the 22q11.2 deletion syndrome].Encephale. 2015 Jun;41(3):266-73. doi: 10.1016/j.encep.2014.10.005. Epub 2014 Dec 16. Encephale. 2015. PMID: 25523123 Review. French.
-
Behavioral and Psychiatric Phenotypes in 22q11.2 Deletion Syndrome.J Dev Behav Pediatr. 2015 Oct;36(8):639-50. doi: 10.1097/DBP.0000000000000210. J Dev Behav Pediatr. 2015. PMID: 26372046 Free PMC article. Review.
Cited by
-
Hyperactive mTORC1 in striatum dysregulates dopamine receptor expression and odor preference behavior.Front Neurosci. 2024 Aug 30;18:1461178. doi: 10.3389/fnins.2024.1461178. eCollection 2024. Front Neurosci. 2024. PMID: 39280263 Free PMC article.
-
The Evolving Role of Animal Models in the Discovery and Development of Novel Treatments for Psychiatric Disorders.Adv Neurobiol. 2023;30:37-99. doi: 10.1007/978-3-031-21054-9_3. Adv Neurobiol. 2023. PMID: 36928846
-
Sleep in 22q11.2 Deletion Syndrome: Current Findings, Challenges, and Future Directions.Curr Psychiatry Rep. 2023 Oct;25(10):479-491. doi: 10.1007/s11920-023-01444-6. Epub 2023 Sep 18. Curr Psychiatry Rep. 2023. PMID: 37721640 Free PMC article. Review.
-
Histological Analysis of a Mouse Model of the 22q11.2 Microdeletion Syndrome.Biomolecules. 2023 Apr 27;13(5):763. doi: 10.3390/biom13050763. Biomolecules. 2023. PMID: 37238632 Free PMC article.
-
Unraveling the enigma of mental disorders: a genetics-first approach and the role of mouse models based on rare disease-susceptible genome variants.Nagoya J Med Sci. 2025 May;87(2):196-210. doi: 10.18999/nagjms.87.2.196. Nagoya J Med Sci. 2025. PMID: 40765798 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

