Farnesoid X receptor via Notch1 directs asymmetric cell division of Sox9+ cells to prevent the development of liver cancer in a mouse model
- PMID: 33845903
- PMCID: PMC8042944
- DOI: 10.1186/s13287-021-02298-6
Farnesoid X receptor via Notch1 directs asymmetric cell division of Sox9+ cells to prevent the development of liver cancer in a mouse model
Abstract
Background: Asymmetrical cell division (ACD) maintains the proper number of stem cells to ensure self-renewal. The rate of symmetric division increases as more cancer stem cells (CSCs) become malignant; however, the signaling pathway network involved in CSC division remains elusive. FXR (Farnesoid X receptor), a ligand-activated transcription factor, has several anti-tumor effects and has been shown to target CSCs. Here, we aimed at evaluating the role of FXR in the regulation of the cell division of CSCs.
Methods: The FXR target gene and downstream molecular mechanisms were confirmed by qRT-PCR, Western blot, luciferase reporter assay, EMAS, Chip, and IF analyses. Pulse-chase BrdU labeling and paired-cell experiments were used to detect the cell division of liver CSCs. Gain- and loss-of-function experiments in Huh7 cells and mouse models were performed to support findings and elucidate the function and underlying mechanisms of FXR-Notch1 in liver CSC division.
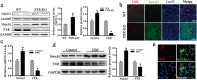
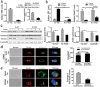
Results: We demonstrated that activation of Notch1 was significantly elevated in the livers of hepatocellular carcinoma (HCC) in Farnesoid X receptor-knockout (FXR-KO) mice and that FXR expression negatively correlated with Notch1 level during chronic liver injury. Activation of FXR induced the asymmetric divisions of Sox9+ liver CSCs and ameliorated liver injury. Mechanistically, FXR directs Sox9+ liver CSCs from symmetry to asymmetry via inhibition of Notch1 expression and activity. Deletion of FXR signaling or over-expression of Notch1 greatly increased Notch1 expression and activity along with ACD reduction. FXR inhibited Notch1 expression by directly binding to its promoter FXRE. FXR also positively regulated Numb expression, contributing to a feedback circuit, which decreased Notch1 activity and directed ACD.
Conclusion: Our findings suggest that FXR represses Notch1 expression and directs ACD of Sox9+ cells to prevent the development of liver cancer.
Keywords: FXR; Liver cancer stem cell; Notch1; Sox9; Symmetric cell division.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Sox9 regulates self-renewal and tumorigenicity by promoting symmetrical cell division of cancer stem cells in hepatocellular carcinoma.Hepatology. 2016 Jul;64(1):117-29. doi: 10.1002/hep.28509. Epub 2016 Mar 25. Hepatology. 2016. PMID: 26910875
-
Upregulation of microRNA-122 by farnesoid X receptor suppresses the growth of hepatocellular carcinoma cells.Mol Cancer. 2015 Aug 25;14:163. doi: 10.1186/s12943-015-0427-9. Mol Cancer. 2015. PMID: 26302777 Free PMC article.
-
Farnesoid X Receptor Attenuates the Tumorigenicity of Liver Cancer Stem Cells by Inhibiting STAT3 Phosphorylation.Int J Mol Sci. 2025 Jan 28;26(3):1122. doi: 10.3390/ijms26031122. Int J Mol Sci. 2025. PMID: 39940889 Free PMC article.
-
Farnesoid X receptor, the bile acid sensing nuclear receptor, in liver regeneration.Acta Pharm Sin B. 2015 Mar;5(2):93-8. doi: 10.1016/j.apsb.2015.01.005. Epub 2015 Feb 20. Acta Pharm Sin B. 2015. PMID: 26579433 Free PMC article. Review.
-
Therapeutic Effectiveness of Anticancer Agents Targeting Different Signaling Molecules Involved in Asymmetric Division of Cancer Stem Cell.Stem Cell Rev Rep. 2023 Jul;19(5):1283-1306. doi: 10.1007/s12015-023-10523-3. Epub 2023 Mar 23. Stem Cell Rev Rep. 2023. PMID: 36952080 Review.
Cited by
-
Crosstalk Between Bile Acids and Intestinal Epithelium: Multidimensional Roles of Farnesoid X Receptor and Takeda G Protein Receptor 5.Int J Mol Sci. 2025 Apr 29;26(9):4240. doi: 10.3390/ijms26094240. Int J Mol Sci. 2025. PMID: 40362481 Free PMC article. Review.
-
Nuclear receptor FXR inhibits ferroptosis to alleviate hepatic ischemia-reperfusion injury by targeting GPX4 in a mouse model.J Mol Histol. 2025 Jul 31;56(4):243. doi: 10.1007/s10735-025-10534-z. J Mol Histol. 2025. PMID: 40742574
-
Metabolic nuclear receptors coordinate energy metabolism to regulate Sox9+ hepatocyte fate.iScience. 2021 Aug 19;24(9):103003. doi: 10.1016/j.isci.2021.103003. eCollection 2021 Sep 24. iScience. 2021. PMID: 34505013 Free PMC article.
-
Inhibition of RFX6 Suppresses the Invasive Ability of Tumor Cells Through the Notch Pathway and Affects Tumor Immunity in Hepatocellular Carcinoma.Front Oncol. 2021 Dec 20;11:801222. doi: 10.3389/fonc.2021.801222. eCollection 2021. Front Oncol. 2021. PMID: 34988028 Free PMC article.
-
Pleiotropic roles of FXR in liver and colorectal cancers.Mol Cell Endocrinol. 2022 Mar 1;543:111543. doi: 10.1016/j.mce.2021.111543. Epub 2022 Jan 4. Mol Cell Endocrinol. 2022. PMID: 34995680 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

